
Cialis ist bekannt für seine lange Wirkdauer von bis zu 36 Stunden. Dadurch unterscheidet es sich deutlich von Viagra. Viele Schweizer vergleichen daher Preise und schauen nach Angeboten unter dem Begriff cialis generika schweiz, da Generika erschwinglicher sind.
Ciml.univ-mrs.fr


This article was originally published in a journal published by
Elsevier, and the attached copy is provided by Elsevier for the
author's benefit and for the benefit of the author's institution, for
non-commercial research and educational use including without
limitation use in instruction at your institution, sending it to specific
colleagues that you know, and providing a copy to your institution's
All other uses, reproduction and distribution, including without
limitation commercial reprints, selling or licensing copies or access,
or posting on open internet sites, your personal or institution's
website or repository, are prohibited. For exceptions, permission
may be sought for such use through Elsevier's permissions site at:

Infection in a dish: high-throughput analyses of bacterialpathogenesisC Le´opold Kurz1,2,3 and Jonathan J Ewbank1,2,3
Diverse aspects of host–pathogen interactions have been
in the infection process regardless of the host. These can
studied using non-mammalian hosts such as Dictyostelium
be identified and characterised using genetically tractable
discoideum, Caenorhabditis elegans, Drosophila melanogaster
and inexpensive non-mammalian models. In addition, the
and Danio rerio for more than 20 years. Over the past two years,
molecular and genetic tools that have been developed for
the use of these model hosts to dissect bacterial virulence
use with these simple organisms, combined with their
mechanisms has been expanded to include the important
well-studied cellular biology and/or immunology, enable
human pathogens Vibrio cholerae and Yersinia pestis.
one to decipher the complex interactions between host
Innovative approaches using these alternative hosts have also
and pathogen.
been developed, enabling the isolation of new antimicrobialsthrough screening large libraries of compounds in a C. elegans
The four organisms listed above have many factors in
Enterococcus faecalis infection model. Host proteins required
common that make them very useful as model hosts, such
by Mycobacterium and Listeria during their invasion and
as the availability of their fully sequenced genomes
intracellular growth have been uncovered using high-
and their ease of culture [1]. These alternative hosts
throughput dsRNA screens in a Drosophila cell culture system,
are being used for approaches as diverse as testing the
and immune evasion mechanisms deployed by Pseudomonas
virulence of chosen pathogen mutants [6,7�], screening
aeruginosa during its infection of flies have been identified.
large banks of pathogen mutants for those with attenu-
Together, these reports further illustrate the potential and
ated virulence [8,9�,10�] or dissecting the host mechan-
relevance of these non-mammalian hosts for modelling many
isms involved in pathogen invasion and intracellular
facets of bacterial infection in mammals.
Addresses1 Centre d'Immunologie de Marseille-Luminy, Universite´ de la
In addition, they have unique features that are relevant to
Me´diterrane´e, Case 906, 13288 Marseille Cedex 9, France
the study of specific aspects of host–pathogen interac-
2 Institut national de la sante´ et de la recherche me´dicale (INSERM),
tions. The amoeba D. discoideum is a professional phago-
U631, 13288 Marseille, France3 Centre National de la Recherche Scientifique (CNRS), UMR6102,
cyte that can be used to decipher the molecular basis of
13288 Marseille, France
phagocytosis and phagosome maturation [4]. Addition-ally, it can give insights into how certain intracellular
Corresponding author: Ewbank, Jonathan J ([email protected])
bacterial pathogens survive in the phagolysosome [14].
The fly D. melanogaster possesses a very well-studied
Current Opinion in Microbiology 2007, 10:10–16
innate immunity [15] that has contributed to the under-standing of immune mechanisms in mammals. More
This review comes from a themed issue on
recently, it has been used to analyze the mechanisms
Host–microbe interactions: bacteria
used by pathogens to evade the host immune system
Edited by Pamela Small and Gisou van der Goot
[16�,17,18��]. Finally, genetic screens for bacterial viru-
Available online 18th December 2006
lence genes in a vertebrate with a fully developedimmune system [19] are possible with the fish D. rerio.
1369-5274/$ – see front matter
This review focuses on recent work with the alternative
# 2006 Elsevier Ltd. All rights reserved.
model hosts D. discoideum, C. elegans, D. melanogaster and
D. rerio in these new investigative paradigms.
New infections modelled with alternative
The use of mamma Author's personal copy
lian models to identify and understand
An increasing number of human bacterial pathogens are
the virulence factors of human pathogens is indispensa-
being tested in non-mammalian hosts in order to con-
ble. Alternative models such as the amoeba Dictyostelium
veniently study their virulence. In addition to established
discoideum, the nematode Caenorhabditis elegans, the insect
models such as Pseudomonas aeruginosa [20,21], Salmonella
Drosophila melanogaster and the fish Danio rerio can be
typhimurium [22–24] or Serratia marcescens [25,26], several
complementary systems for such studies [1–5]. This is
pathogens including Listeria monocytogenes [27,28], Yersi-
possible because many human pathogens have a low
nia pestis (see below) and Vibrio cholerae, the causal agent
species-specificity and can infect hosts ranging from
of cholera, have recently been added to the list of micro-
insects and nematodes to fish, as well as other mammals.
organisms that are capable of causing lethal infection of
They rely on universal virulence factors that are involved
the nematode and the fly.
Current Opinion in Microbiology 2007, 10:10–16
Infection in a dish: high-throughput analyses of bacterial pathogenesis Kurz and Ewbank
In humans, expression of cholera toxin (CT) by V. cholerae
to identify in vivo new antibacterial molecules. A similar
provokes a rise in cAMP in the intestinal epithelium, the
system involving flies to be used to identify antifungal
opening of ion channels and consequently, loss of water
drugs is also being developed [31,32].
into the intestinal lumen. In mice, this secretory diarrhoeacan be successfully treated with the channel-blocker
Random screens for the identification of
clotrimazole. It has now been reported that oral infection
bacterial virulence genes
of the fruit fly by V. cholerae leads to the death of the
Three recent reports [8,9�,10�] using D. discoideum, C.
animals in a manner somewhat similar to that observed in
elegans and D. rerio as hosts to screen bacterial mutant
humans, including rapid weight-loss [7�]. CT is required
libraries of Klebsiella pneumoniae, Y. pestis and Streptococcus
for full virulence in the fly model and, remarkably, flies
iniae, respectively, have further strengthened the rele-
with loss-of-function mutations in genes encoding homo-
vance of these simple hosts.
logues of the known targets of CT resist infection.
Furthermore, clotrimazole can help cure flies infected
K. pneumoniae is an important human pathogen that, as its
with V. cholerae [7�].
name suggests, causes pneumonia. Its interaction withalveolar macrophages can be modelled using D. discoideum
During the lethal colonization of the C. elegans intestine
as a surrogate phagocyte. D. discoideum is normally able to
by V. cholerae, however, CT does not appear to play an
feed on wild type Klebsiella. Cosson and colleagues
important role [6]. But, using a reverse genetic approach,
[10�,33] elegantly combined the genetics of D. discoideum
Vaitkevicius et al. [6] demonstrated that the quorum
and the genetics of K. pneumoniae. They first identified a
sensing regulated protease PrtV is essential for this kill-
new gene ( phg1) that, when mutated, rendered the
ing. Moreover, they obtained data suggesting that this
amoeba especially susceptible to infection and unable
protease is important to V. cholerae in its natural niche [29]
to grow on Klebsiella. They then isolated Klebsiella
for its resistance to the marine plankton that graze on the
mutants that supported the growth of the phg1 mutant
bacterium. Finally, they measured an increased interleu-
amoeba: among the mutated bacterial genes were several
kin-8 (IL–8) secretion in human epithelial intestinal cells
that were required for biosynthesis of lipopolysaccharides
exposed to a V. cholerae prtV deletion mutant, compared to
and amino acids. They tested several of the isolated
that of the parental strain, suggesting a role for this
bacterial mutants in a mouse pneumonia model and found
protease in modulating (directly or indirectly) the host
an attenuation of virulence [10�].
response in vertebrates [6].
The genetic manipulation of both host and pathogen
Together, these reports illustrate to what extent nema-
enabled the authors to create a 2D virulence array show-
tode and fly can be relevant for the study of the causative
ing that distinct groups of host genes are necessary to
agent of cholera. Importantly, the work by Blow et al.
resist infection by various bacterial pathogens and
[7�] are compatible with the idea of using Drosophila to
mutants (Figure 2). They were also able to demonstrate
screen for chemicals that inhibit CT in vivo, following
conservation of both virulence factors and defence genes
a precedent set by the Ausubel laboratory [30��], using
because Drosophila phg1 mutants are more susceptible to
K. pneumoniae infection [10�].
In vivo screens for new antimicrobials
Y. pestis, the causative agent of plague, can form a biofilm
The massive use of antibiotics, combined with the high
that is important for dissemination by its vector, the
adaptation capacity of bacteria has created a huge public
flea. A Y. pestis biofilm can also accumulate on the head
health problem with many human pathogens becoming
of C. elegans, and this is clearly a more accessible model
resistant to multiple antibiotics. Therefore, there is a real
for studying biofilm function than is looking in the gut
need for new antibiotic molecules. Moy et al. [30��]
of the flea [34]. As biofilm formation is only one aspect of
cleverly used an infection system involving a C. elegans
Y. pestis pathogenicity, Styer et al. [9�] developed a
immunocompromised mutant and Enterococcus faecalis to
nematode-based infection system to identify Y. pestis
screen thousands of synthetic and natural molecules to
virulence genes not related to biofilm formation. They
Author's personal copy
promoted host survival (Figure 1). This in
showed that a biofilm-deficient mutant of Y. pestis colo-
vivo screen not only permitted the identification of eight
nises the intestine of C. elegans and provokes an early
molecules that affect bacterial growth in vitro (minimum
death of the host. They used this infection model to
inhibitory concentration [MIC] <35 mg ml 1) but also of
screen a bank of Y. pestis mutants for those with attenu-
eight other products that either impair pathogen viru-
ated virulence in the nematode. Remarkably, despite the
lence or boost host innate immunity in the absence of
differences between nematodes and mammals, they
significant in vitro activity (MIC > 125 mg ml 1) [30��].
identified two genes necessary for full virulence in an
Even though the efficiency and toxicity of the identified
intranasal mouse model of Y. pestis pathogenesis, genes
molecules does need to be tested in mammals, this
that had previously not been implicated in Y. pestis
system represents a very promising screening platform
pathogenicity [9�].
Current Opinion in Microbiology 2007, 10:10–16
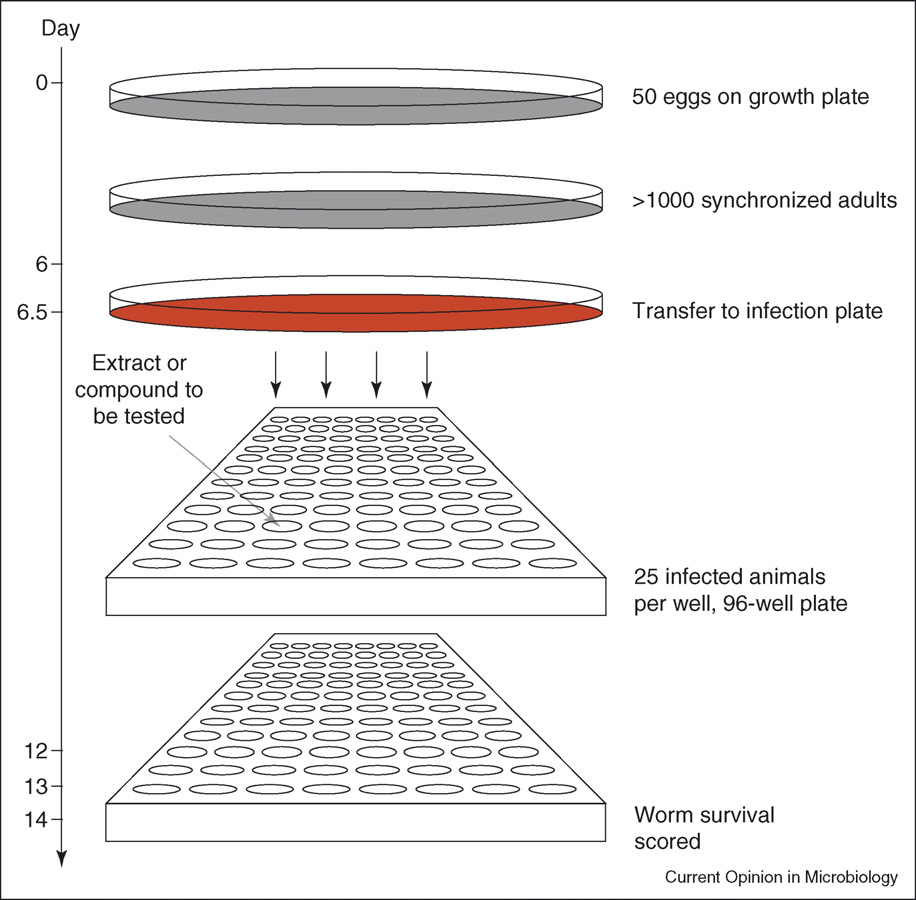
Host–microbe interactions: bacteria
Protocol used by Moy et al. [30��] to screen in vivo for new antimicrobial compounds using an established C. elegans–E. faecalis infection system.
After culture and amplification of nematode numbers on growth plates (seeded with the Escherichia coli strain OP50) synchronised populationsof worms are transferred to infection plates, seeded with E. faecalis strain MMH594. After 8 h, worms are washed off the plates and approximately25 individuals added to each well of a 96-well microtitre plate and then assayed for their survival. Compounds or extracts that extended wormsurvival by twofold to threefold after 6–8 day's culture were selected for further analyses. Whereas this screen was carried out manually, automationof different steps is possible with tools such as Union Biometrica Biosort (http://www.unionbio.com/products/copas2.html). It is important to notethat similar screens for antimicrobial compounds can be designed using C. elegans and other pathogens. The time when worm survival is scoredwill vary depending on the pathogen used.
S. iniae is a bacterial pathogen that is able to infect fish
involved in invasion and survival in human macrophages
and humans. To analyze the interaction between strep-
tococcal pathogens and their natural hosts, Miller et al.
[8] created a bank of bacterial mutants and screened it
These three studies further validate the use of non-mam-
using zebrafish. They wished to identify bacterial
malian hosts for large-scale screens to identify bacterial
mutants specifically deficient in their capacity to disse-
virulence genes relevant to infection in mammals. More-
minate in the brain. To facilitate the screening process,
over, the genetic manipulation of the host, as exemplified
they used a signature-tagged mutagenesis strategy
by the work of Benghezal et al. [10�], expands the range of
Author's personal copy
rmitted the analysis of fish co-infected by
models available for this kind of screening approach, in a
a pool of 12 distinct mutant strains. Doing so, they
manner reminiscent of the directed modification of mice,
screened 1128 signature-tagged transposon bacterial
through trangenesis [36] or the creation of human-mouse
mutants and determined which bacterial mutants were
chimeras [37], but without any of the ethical concerns.
not present in brain extracts from infected fish. Interest-ingly, 7 out of the 41 bacterial mutants isolated had
Identification of host molecules required for
transposon insertions in genes required for the production
pathogenesis and how the pathogen evades
of capsular polysaccharides. Finally, using the bacterial
the immune system
mutants they isolated, they showed in a human whole
The host factors involved in the infection processes are
blood assay for phagocytosis that the capsule of S. iniae is
not restricted to ‘immunity genes' such as those coding
Current Opinion in Microbiology 2007, 10:10–16
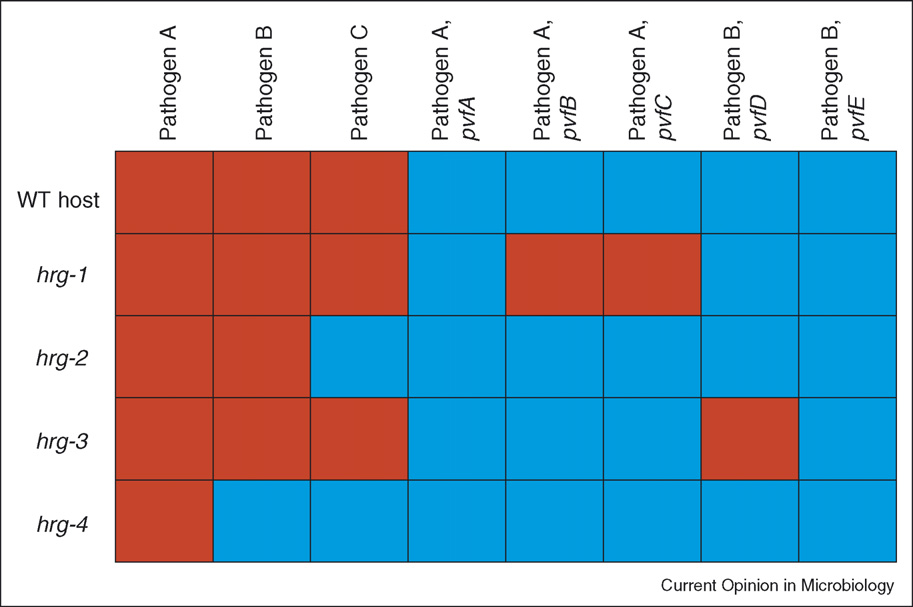
Infection in a dish: high-throughput analyses of bacterial pathogenesis Kurz and Ewbank
Hypothetical host–pathogen 2D array inspired by data from Benghezal et al. [10�]. The ability of host mutants to resist (blue) or their susceptibilityto (red) different bacterial strains and bacterial mutants is indicated. Gene names are arbitrary with hrg and pvf for ‘host resistance gene' and‘pathogen virulence factor', respectively. Based on this matrix, it can be speculated that hrg-1 is specifically involved in a mechanism necessaryfor host resistance to bacterial virulence factors encoded by pvfB and pvfC. The hrg-3–pvfD interaction would correspond to the case describedby Liehl et al. [18��] with hrg-3 and pvfD being the Drosophila Imd and Pseudomonas aprA genes, respectively. Finally, hrg-2 and hrg-4 couldencode host proteins necessary for bacterial invasion by pathogen C and pathogens B and C, respectively, corresponding to the observationsdescribed in the reports by Philips et al. [12�] and Agaisse et al. [13�].
for interleukins or Toll-like receptors (TLRs). This is
family. This work also highlighted a role for autophagy in
especially the case for intracellular bacterial pathogens
the control of L. monocytogenes infection [12�].
that have to enter the cell and avoid being degraded inphagolysosomes. Therefore, intracellular bacteria have
In contrast to these two studies, which used automated
developed many ways to hijack the endocytic or phago-
microscopy, a third study was performed manually [11��].
cytic routes [38,39]. Macrophages are often confronted by
In this painstaking project, interest was focused espe-
intracellular pathogens because they are professional
cially on the interaction between the L. monocytogenes
phagocytes. The Drosophila S2 macrophage-like cell line
toxin listeriolysin O (LLO) and host factors that enable
has now been used in three large-scale RNA interference
the bacteria to escape from the phagosome. The authors
(RNAi) screens in order to identify host factors required
used RNAi to inactivate host genes and combined this
for entry and survival of intracellular bacterial pathogens
with bacteria mutated in LLO. In a first set of experi-
[11��,12�,13�]. The first two analyses combined auto-
ments, they used an LLO-deficient bacterial strain
mated microscopy with the use of green fluorescent
unable to leave the phagosomes of normal cells and
protein (GFP)-tagged Mycobacterium fortuitum [13�] or
screened for dsRNAs that restored the capacity of these
Listeria monocytogenes [12�] to screen a bank containing
mutants to escape into the cytoplasm. The corresponding
21 300 dsRNAs (targeting >95% of annotated Drosophila
genes would be expected to be elements of the host
genes in a redundant fashion). They showed that factors
pathways targeted by LLO. In a second set of experi-
involved in vesi Author's personal copy
cle trafficking and actin cytoskeleton
ments, they used a bacterial mutant producing a LLO
organization are necessary for internalization and intra-
toxin that lacks the PEST sequence which normally
cellular survival of these two pathogens. Moreover, they
makes the protein relatively unstable. They screened
identified Peste (French for ‘plague'), a Drosophila homo-
for dsRNAs that rendered S2 cells more susceptible to
logue of the scavenger receptor CD36, as being crucial for
this stable toxin in order to determine which host
entry of L. monocytogenes and M. fortuitum into the S2 cells,
enzymes control LLO toxicity. On the basis of their
whilst being dispensable for phagocytosis in general
results, they proposed a model in which the pore-forming
[12�,13�]. On the basis of these observations, the study
LLO inserts into the membrane of the L. monocytogenes-
was extended to mammalian cells and new roles in uptake
containing phagosome, thus impairing its acidification
of bacteria were described for members of the CD36
and maturation. Concerning the host's control of LLO
Current Opinion in Microbiology 2007, 10:10–16
Host–microbe interactions: bacteria
toxicity, their screen identified serine palmitoyl-CoA
bacterial pathogens for their mammalian host. For
transferase (SPT), which is an enzyme necessary for
instance, some virulence genes involved in mammalian
sphingolipid metabolism, as a key factor for host resis-
pathogenesis are only expressed at 37 8C, whereas not all
tance [11��].
the model animals described in this review can be grownat this temperature [5]. Moreover, C. elegans does not
The experimental systems described in these three reports
possess macrophage-like cells [42] and some receptors
can thus be used to shed light on the complex interactions
necessary for the engulfment of intracellular bacterial
between the host and an intracellular pathogen that are
pathogens in mammalian cells are absent from the surface
both fighting for their survival. But just as is the case for any
of non-mammalian cells, thus limiting the utility of sim-
model system, the results come with several caveats. It is
ple organisms for the study of intracellular pathogenesis.
well known that a dsRNA can interfere with off-target
The same is true concerning mammalian signalling path-
genes and so generate false positive results [40]. Conver-
ways specifically targeted and hijacked by some patho-
sely, important genes can be missed if they are not
gens (e.g. although it possesses one TLR, NF-kB
expressed in or on S2 cells, as is indeed the case for some
transcription factors, crucial for mammalian immunity,
receptors involved in phagocytosis (Istvan Ando and Dan
are not present in C. elegans [43]).
Hultmark, personal communication). Nevertheless, in thelong term, by combining large-scale screens in the host and
the pathogen, it will be possible to define a host–pathogen
Although evolutionary divergence from mammals can
interactome (Figure 2) [41].
limit the pertinence of simple model animals, the papersdescribed in this review demonstrate that there is a
Extracellular bacterial pathogens are usually not able to
wide-spread conservation of host–pathogen interactions
survive phagocytosis. Many, however, have developed
at the molecular and physiological levels. In the light of
strategies to counteract the humoral arm of the host
this, the phylogenetic distance between a model system
immune system. A handful of recent articles have demon-
and mammals can even be considered a boon because
strated that infection of D. melanogaster with Pseudomonas
conserved interactions are frequently the most impor-
is a most suitable system to study the host immune
tant. Therefore, given the practical advantages asso-
response and to uncover the strategies used by the
ciated with their use, non-mammalian models are
pathogen to elude defence mechanisms. In one article
increasingly being recognized as attractive alternatives
[17], the role of the Pseudomonas exotoxin ExoS was
to more traditional models [5]. Moreover, it is probable
directly addressed by expressing this toxin either ectopi-
that many of the virulence mechanisms that pathogens
cally in the eye or ubiquitously throughout the fly. The
use during their infection of humans in fact evolved
authors showed with these transgenic systems that ExoS
because they confer a survival advantage in the natural
inhibits the activity of a host Rho GTPase in vivo and that
ecological niche, and so are best studied using their
ubiquitous ExoS expression impairs the phagocytic capa-
natural predators, such as D. discoideum and C. elegans.
city of fly macrophages without affecting induction of
After a period when these model systems were used in
antimicrobial peptide genes [17]. In a complementary
essentially one-sided approaches (e.g. screening banks of
study, Liehl et al. [18��] used host and pathogen mutants
bacterial mutants for virulence genes or identifying the
to demonstrate that the Pseudomonas AprA metallopro-
host targets of bacterial virulence factors), more and
tease directly degrades fly antimicrobial peptides. This
more studies are now exploiting a combination of bac-
protease thereby acts as a virulence factor by enhancing
terial and host genetics to address the molecular basis of
bacterial survival within the host body fluid. In addition to
pathogenicity and defence. The future promises to
these reports, Apidianakis et al. [16�] compared microarray
reveal details of the intimate but deadly dance between
results from flies infected by virulent or avirulent P.
pathogen and host that has been going on since the birth
aeruginosa strains. Strikingly, this analysis revealed an
of eukaryotes.
as yet uncharacterised mechanism used by P. aeruginosain the early phases of the infection to limit expression of
Drosophila antimicrobial genes at the transcriptional level.
It has recently been shown using C. elegans, the P. aeru-ginosa strain PA14 and a PA14 gacA mutant (that is highly
Author's personal copy
studies illustrate the potential use of
attenuated) that the intrinsic virulence of PA14 is a major
genetically tractable non-mammalian hosts, with charac-
elicitor of the host's innate immune response [44�]. In
terized immune systems, to decipher the mechanisms
addition, by using the same animal model and by com-
pathogens employ to evade host defenses. As exemplified
paring the genome of the P. aeruginosa strain PA14 with
above, it is possible to have a global approach and/or to
that of PA01, it has been demonstrated that Pseudomonas
precisely address the role of a specific bacterial protein.
virulence is multifactorial and necessitates the combina-torial action of multiple virulence factors that interact
The principal drawbacks with these models are associated
in a distinct manner, depending on the bacterial genetic
with bacterial physiology and the specificity of certain
background [45�].
Current Opinion in Microbiology 2007, 10:10–16
Infection in a dish: high-throughput analyses of bacterial pathogenesis Kurz and Ewbank
12. Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N,�
Higgins DE: Genome-wide RNAi screen for host factors
We thank Pierre Golstein for helpful criticism. Work in the authors'
required for intracellular bacterial infection. Science 2005,
laboratory is supported by the Fondation Recherche Me´dicale, INSERM,
the CNRS, the French Ministry of Research, Marseille-Nice Ge´nopole,
See annotation for [13�].
the Re´seau Nationale des Ge´nopoles, the European Union and theFrench National Research Agency (ANR).
13. Philips JA, Rubin EJ, Perrimon N: Drosophila RNAi screen�
reveals CD36 family member required for mycobacterialinfection. Science 2005, 309:1251-1253.
References and recommended reading
To uncover host factors necessary for intracellular bacterial infection,
Papers of particular interest, published within the period of review,
these two papers used a genome-wide RNAi bank, a Drosophila macro-
have been highlighted as:
phage-like cell line, automated microscopy and GFP-tagged L. mono-cytogenes [12�] or M. fortuitum [13�]. Several hundred dsRNA that altered
� of special interest
the progression of infection were identified. The results obtained in the
�� of outstanding interest
two screens were directly compared [12�]. Peste, a member of the CD36family was found to be crucial for the entry of both bacteria into cells.
Steinert M, Heuner K: Dictyostelium as host model for
14. Li Z, Solomon JM, Isberg RR: Dictyostelium discoideum strains
lacking the RtoA protein are defective for maturation of the
pathogenesis. Cell Microbiol 2005, 7:307-314.
Legionella pneumophila replication vacuole. Cell Microbiol
Sifri CD, Begun J, Ausubel FM: The worm has turned–microbial
2005, 7:431-442.
virulence modeled in Caenorhabditis elegans. Trends Microbiol2005, 13:119-127.
15. Royet J, Reichhart JM, Hoffmann JA: Sensing and signaling
during infection in Drosophila. Curr Opin Immunol 2005,
Vodovar N, Acosta C, Lemaitre B, Boccard F: Drosophila: a
polyvalent model to decipher host–pathogen interactions.
Trends Microbiol 2004, 12:235-242.
16. Apidianakis Y, Mindrinos MN, Xiao W, Lau GW, Baldini RL,�
Davis RW, Rahme LG: Profiling early infection responses:
van der Sar AM, Musters RJ, van Eeden FJ, Appelmelk BJ,
Pseudomonas aeruginosa eludes host defenses by
Vandenbroucke-Grauls CM, Bitter W: Zebrafish embryos as a
suppressing antimicrobial peptide gene expression. Proc Natl
model host for the real time analysis of Salmonella
Acad Sci USA 2005, 102:2573-2578.
typhimurium infections. Cell Microbiol 2003, 5:601-611.
In this study, the authors performed microarray analyses on flies infectedwith either wild type P. aeruginosa or an avirulent mutant. Strikingly, they
Pradel E, Ewbank JJ: Genetic models in pathogenesis.
uncovered an enigmatic mechanism used by P. aeruginosa specifically to
Annu Rev Genet 2004, 38:347-363.
silence the expression of genes encoding antimicrobial proteins.
Vaitkevicius K, Lindmark B, Ou G, Song T, Toma C, Iwanaga M,
17. Avet-Rochex A, Bergeret E, Attree I, Meister M, Fauvarque MO:
Zhu J, Andersson A, Hammarstrom ML, Tuck S et al.: A Vibrio
Suppression of Drosophila cellular immunity by directed
cholerae protease needed for killing of Caenorhabditis
expression of the ExoS toxin GAP domain of Pseudomonas
elegans has a role in protection from natural predator grazing.
aeruginosa. Cell Microbiol 2005, 7:799-810.
Proc Natl Acad Sci USA 2006, 103:9280-9285.
18. Liehl P, Blight M, Vodovar N, Boccard F, Lemaitre B: Prevalence
Blow NS, Salomon RN, Garrity K, Reveillaud I, Kopin A,
of local immune response against oral infection in a
Jackson FR, Watnick PI: Vibrio cholerae infection of Drosophila
Drosophila/Pseudomonas infection model. PLoS Pathog 2006,
melanogaster mimics the human disease cholera.
PLoS Pathog 2005, 1:e8.
The authors developed an oral infection model between Drosophila and a
This report establishes D. melanogaster as a new in vivo model with which
newly identified entomopathogen Pseudomonas entomophila and iden-
to study V. cholerae infection. As for mammals, the infection is per os, and
tified and characterised a bacterial metalloprotease abrogating host
CT is essential. Moreover, the host machinery involved in the subsequent
defences through degradation of antimicrobial peptides.
secretory diarrhoea is conserved.
19. Trede NS, Langenau DM, Traver D, Look AT, Zon LI: The use of
Miller JD, Neely MN: Large-scale screen highlights the
zebrafish to understand immunity. Immunity 2004, 20:367-379.
importance of capsule for virulence in the zoonotic pathogenStreptococcus iniae. Infect Immun 2005, 73:921-934.
20. D'Argenio DA, Gallagher LA, Berg CA, Manoil C: Drosophila as a
model host for Pseudomonas aeruginosa infection. J Bacteriol
Styer KL, Hopkins GW, Bartra SS, Plano GV, Frothingham R,
Aballay A: Yersinia pestis kills Caenorhabditis elegans by abiofilm-independent process that involves novel virulence
21. Tan MW, Mahajan-Miklos S, Ausubel FM: Killing of
factors. EMBO Rep 2005, 6:992-997.
Caenorhabditis elegans by Pseudomonas aeruginosa used to
The authors demonstrate that Y. pestis can infect C. elegans in a biofilm-
model mammalian bacterial pathogenesis. Proc Natl Acad Sci
independent fashion and they use this infection model to identify new Y.
USA 1999, 96:715-720.
pestis factors that are also necessary for full virulence in mice.
22. Brandt SM, Dionne MS, Khush RS, Pham LN, Vigdal TJ,
10. Benghezal M, Fauvarque MO, Tournebize R, Froquet R,
Schneider DS: Secreted bacterial effectors and host-produced
Marchetti A, Bergeret E, Lardy B, Klein G, Sansonetti P,
Eiger/TNF drive death in a Salmonella-infected fruit fly.
Charette SJ et al.: Specific host genes required for the killing
PLoS Biol 2004, 2:e418.
of Klebsiella bacteria by phagocytes. Cell Microbiol 2006,8:139-148.
23. Aballay A, Yorgey P, Ausubel FM: Salmonella typhimurium
In this study, D. discoideum and K. pneumoniae genetics were combined
proliferates and establishes a persistent infection in the
to identify host resistance genes and to screen for pathogen virulence
intestine of Caenorhabditis elegans. Curr Biol 2000,
factors, respectively. Importantly, the bacterial virulence genes identified
are necessary for
24. Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ:
Caenorhabditis elegans is a model host for Salmonella
Portnoy DA: Use Author's personal copy
full-killing in a mouse pneumonia model.
JP, Stuurman N, Wiedemann U, Vale RD,
of RNA interference in Drosophila S2 cells to
typhimurium. Curr Biol 2000, 10:1543-1545.
identify host pathways controlling compartmentalization ofan intracellular pathogen. Proc Natl Acad Sci USA 2005,
25. Flyg C, Kenne K, Boman HG: Insect pathogenic properties of
Serratia marcescens: phage-resistant mutants with a
The authors used a bank of 7216 dsRNA to identify host factors involved
decreased resistance to Cecropia immunity and a decreased
in pathogenesis of Drosophila S2 cells infected by L. monocytogenes. To
virulence to Drosophila. J Gen Microbiol 1980, 120:173-181.
uncover the host pathways targeted by the bacterial LLO toxin and thehost mechanisms protecting the cell from this toxin, they combined this
26. Kurz CL, Chauvet S, Andres E, Aurouze M, Vallet I, Michel GP,
large-scale approach with bacterial mutants. They identified host proteins
Uh M, Celli J, Filloux A, De Bentzmann S et al.: Virulence factors
not previously involved in Listeria pathogenesis and enzymes involved in
of the human opportunistic pathogen Serratia marcescens
identified by in vivo screening. EMBO J 2003, 22:1451-1460.
Current Opinion in Microbiology 2007, 10:10–16
Host–microbe interactions: bacteria
27. Thomsen LE, Slutz SS, Tan MW, Ingmer H: Caenorhabditis
37. Legrand N, Weijer K, Spits H: Experimental models to study
elegans is a model host for Listeria monocytogenes.
development and function of the human immune system
Appl Environ Microbiol 2006, 72:1700-1701.
in vivo. J Immunol 2006, 176:2053-2058.
28. Mansfield BE, Dionne MS, Schneider DS, Freitag NE:
38. Henry T, Gorvel JP, Meresse S: Molecular motors hijacking by
Exploration of host–pathogen interactions using Listeria
intracellular pathogens. Cell Microbiol 2006, 8:23-32.
monocytogenes and Drosophila melanogaster. Cell Microbiol2003, 5:901-911.
39. Ismail N, Olano JP, Feng HM, Walker DH: Current status of
immune mechanisms of killing of intracellular
29. Reidl J, Klose KE: Vibrio cholerae and cholera: out of the water
microorganisms. FEMS Microbiol Lett 2002, 207:111-120.
and into the host. FEMS Microbiol Rev 2002, 26:125-139.
40. Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P,
30. Moy TI, Ball AR, Anklesaria Z, Casadei G, Lewis K, Ausubel FM:
Perrimon N, Mathey-Prevot B: Evidence of off-target effects
Identification of novel antimicrobials using a live-animal
associated with long dsRNAs in Drosophila melanogaster
infection model. Proc Natl Acad Sci USA 2006, 103:10414-10419.
cell-based assays. Nat Methods 2006, 3:833-838.
Using a C. elegans–E. faecalis infection model, the authors developed ahigh-throughput platform to identify antimicrobial compounds in vivo.
41. Tenor JL, McCormick BA, Ausubel FM, Aballay A: Caenorhabditis
Interestingly, the majority of the identified molecules are not antimicrobial
elegans-based screen identifies Salmonella virulence factors
in vitro, but only efficient in vivo. This suggests that they might either boost
required for conserved host-pathogen interactions. Curr Biol
the host immune system or act through the neutralization of some
2004, 14:1018-1024.
bacterial virulence factors.
42. Ewbank JJ: Tackling both sides of the host–pathogen equation
31. Lionakis MS, Kontoyiannis DP: Fruit flies as a minihost model
with Caenorhabditis elegans. Microbes Infect 2002, 4:247-256.
for studying drug activity and virulence in Aspergillus.
Med Mycol 2005, 43(Suppl 1):S111-S114.
43. Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP,
Solari R, Johnson CD, Ewbank JJ: A reverse genetic analysis of
32. Chamilos G, Lionakis MS, Lewis RE, Lopez-Ribot JL, Saville SP,
components of the Toll signalling pathway in Caenorhabditis
Albert ND, Halder G, Kontoyiannis DP: Drosophila melanogaster
elegans. Curr Biol 2001, 11:809-821.
as a facile model for large-scale studies of virulencemechanisms and antifungal drug efficacy in Candida species.
44. Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, Kim DH:
J Infect Dis 2006, 193:1014-1022.
p38 MAPK regulates expression of immune response genesand contributes to longevity in C. elegans. PLoS Genetics 2006,
33. Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E,
Letourneur F, Bruckert F, Cosson P: Phg1p is a nine-
Using the well-characterized C. elegans-PA14 infection model and a
transmembrane protein superfamily member involved in
number of worm and bacteria mutants, the authors performed series
Dictyostelium adhesion and phagocytosis. J Biol Chem 2000,
of microarray analyses to elucidate the host pathways involved in the
innate immune response. They suggest that virulence factors might
34. Darby C, Hsu JW, Ghori N, Falkow S: Caenorhabditis elegans:
trigger the nematode's defences.
plague bacteria biofilm blocks food intake. Nature 2002,
45. Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S,
Diggins LT, He J, Saucier M, Deziel E et al.: Genomic analysis
35. Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW:
reveals that Pseudomonas aeruginosa virulence is
Simultaneous identification of bacterial virulence genes by
combinatorial. Genome Biol 2006, 7:R90.
negative selection. Science 1995, 269:400-403.
While the genome of the P. aeruginosa strains PA14 and PA01 areremarkably similar, their pathogenicity against C. elegans is completely
36. Lecuit M, Cossart P: Genetically-modified-animal models for
different. Using the nematode as a model host, the authors determined
human infections: the Listeria paradigm. Trends Mol Med 2002,
that genes required for pathogenicity in a given P. aeruginosa strain are
neither required for nor predictive of virulence in other strains.
Author's personal copy
Current Opinion in Microbiology 2007, 10:10–16
Source: http://www.ciml.univ-mrs.fr/EWBANK_jonathan/Articles/Kurz_CurrOpMicro.pdf
The antimalarial and people suffering from the disease per year. Africa accounts forover 90% of reported cases, with an annual 20% increase of cytotoxic drug cryptolepine malaria-related illness and death. Malaria is responsible for as many deaths per annum as AIDS for all of the last 15 years. Drug intercalates into DNA at resistance to malaria has become one of the most significantthreats to human health and the search for new effective drugs is
FULL PAPER International Journal of Recent Trends in Engineering, Vol 2, No. 1, November 2009 EVISTA – Interactive Visual Clustering System K. Thangavel1, P. Alagambigai2 1 Department of Computer Science, Periyar University, Salem, Tamilnadu, India Email: [email protected] 2 Department of Computer Applications, Easwari Engineering College, Chennai, Tamilnadu, India









