
Cialis ist bekannt für seine lange Wirkdauer von bis zu 36 Stunden. Dadurch unterscheidet es sich deutlich von Viagra. Viele Schweizer vergleichen daher Preise und schauen nach Angeboten unter dem Begriff cialis generika schweiz, da Generika erschwinglicher sind.
200913660 8605.8610

Phenothiazines inhibit S100A4 function
by inducing protein oligomerizationVladimir N. Malashkevicha, Natalya G. Dulyaninovaa, Udupi A. Ramagopala, Melissa A. Lirianob, Kristen M. Varneyb,David Knighta2, Michael Brenowitza, David J. Weberb, Steven C. Almoa, and Anne R. Bresnicka,1
aDepartment of Biochemistry, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461; and bDepartment of Biochemistry and
Molecular Biology, University of Maryland School of Medicine, 108 North Greene Street, Baltimore, MD, 21201
Edited by Gregory A. Petsko, Brandeis University, Waltham, MA, and approved April 1, 2010 (received for review November 25, 2009)
S100A4, a member of the S100 family of Ca2þ-binding proteins,
rheumatoid arthritis, cardiac hypertrophy, and kidney fibrosis
regulates carcinoma cell motility via interactions with myosin-
(9 and 10). Given the contribution of S100A4 activity to a variety
IIA. Numerous studies indicate that S100A4 is not simply a marker
of human pathologies, it has received significant attention as a
for metastatic disease, but rather has a direct role in metastatic pro-
possible target for therapeutic intervention.
gression. These observations suggest that S100A4 is an excellent
The disruption of S100A4 binding to its protein targets
target for therapeutic intervention. Using a unique biosensor-
provides the most straightforward means for inhibiting S100A4
based assay, trifluoperazine (TFP) was identified as an inhibitor
activity. S100A4, like other S100 family members, is reported
that disrupts the S100A4/myosin-IIA interaction. To examine the
to have multiple Ca2þ-dependent protein targets that include
interaction of S100A4 with TFP, we determined the 2.3 Å crystal
the cytoskeletal proteins nonmuscle myosin-IIA, tropomyosin,
structure of human Ca2þ-S100A4 bound to TFP. Two TFP molecules
and F-actin (11–13), signaling proteins such as liprin β1 (14),
bind within the hydrophobic target binding pocket of Ca2þ-S100A4
the transcription factor p53 (15) and cell surface molecules
with no significant conformational changes observed in the protein
annexin A2 and Tag7 (16 and 17). At this time there is little struc-
upon complex formation. NMR chemical shift perturbations are
tural information on S100A4-target complexes (5), which will be
consistent with the crystal structure and demonstrate that TFP
needed for the development of S100A4-based therapies.
binds to the target binding cleft of S100A4 in solution. Remarkably,
Using a biosensor that reports on the Ca2þ-activation status of
TFP binding results in the assembly of five Ca2þ-S100A4/TFP dimers
S100A4, we previously identified several phenothiazines that
into a tightly packed pentameric ring. Within each pentamer most
block the Ca2þ-induced fluorescence increase of the biosensor
of the contacts between S100A4 dimers occurs through the TFP
in the low to midmicromolar range (18). Several phenothiazines,
moieties. The Ca2þ-S100A4/prochlorperazine (PCP) complex exhi-
including trifluoperazine (TFP), block the ability of S100A4 to
bits a similar pentameric assembly. Equilibrium sedimentation
depolymerize myosin-IIA filaments. To examine the mechanism
and cross-linking studies demonstrate the cooperative formation
of TFP inhibition of S100A4, we determined the X-ray structure
of a similarly sized S100A4/TFP oligomer in solution. Assays exam-
of the Ca2þ-S100A4/TFP complex. The structure shows that TFP
ining the ability of TFP to block S100A4-mediated disassembly of
binding results in the assembly of five Ca2þ-S100A4/TFP dimers
myosin-IIA filaments demonstrate that significant inhibition of
into a tightly packed pentameric ring via interactions between the
S100A4 function occurs only at TFP concentrations that promote
two TFP molecules located in the hydrophobic target binding
S100A4 oligomerization. Together these studies support a unique
pocket of Ca2þ-S100A4. PCP induces a similar pentameric
mode of inhibition in which phenothiazines disrupt the S100A4/
assembly, suggesting a general mechanism for phenothiazine-
myosin-IIA interaction by sequestering S100A4 via small mole-
mediated oligomerization. Biochemical and biophysical assays
support the formation of a similarly sized Ca2þ-S100A4/TFP
oligomer in solution, and demonstrate that significant inhibition
calcium ∣ X-ray crystallography ∣ NMR ∣ small molecule inhibitor ∣ metastasis
of S100A4 activity occurs only at TFP concentrations that pro-
mote S100A4 oligomerization. We propose a unique mode of
he S100 proteins, of which there are more than 20 members,
inhibition in which phenothiazines disrupt S100A4 activity via
are characterized by their solubility in 100% saturated ammo-
small molecule-induced oligomerization.
nium sulfate (1 and 2). Each S100 family member contains two
Ca2þ-binding loops; a C-terminal "typical" EF-hand comprised of
12 residues and an N-terminal pseudo EF-hand consisting of 14
Structure Determination. A molecular replacement search with the
residues. The basic organization of the S100 proteins is a sym-
Ca2þ-activated human S100A4 dimeric structure (PDB 2Q91) (5)
metric, antiparallel homodimer, in which the N- and C- terminal
produced unique rotation and translation solutions correspond-
helices (helices 1 and 4) from each subunit interact to form a
ing to 10 S100A4 dimers in the asymmetric unit. In the final mod-
stable four helix bundle that serves as the dimer interface.
el, 95.8%, 3.5%, and 0.7% of residues are in favorable, allowed,
Calcium binding to the C-terminal typical EF-hand significantly
alters the angle between helices 3 and 4, which flank the C-term-
inal Ca2þ-binding loop, and exposes a hydrophobic cleft that
Author contributions: N.G.D., D.J.W., S.C.A., and A.R.B. designed research; V.M., N.G.D.,
constitutes a binding surface for target proteins (3–5). Thus
U.A.R., M.A.L., K.M.V., D.K., M.B., and A.R.B. performed research; V.M., N.G.D., U.A.R.,M.A.L., K.M.V., M.B., D.J.W., S.C.A., and A.R.B. analyzed data; and V.M., M.B., D.J.W.,
the S100 proteins operate as calcium-activated switches that bind
S.C.A., and A.R.B. wrote the paper.
and regulate the activity of diverse protein targets.
The authors declare no conflict of interest.
S100 proteins are expressed in a tissue and cell specific manner.
This article is a PNAS Direct Submission.
Elevated expression of individual family members is associated
Data deposition: The sequences reported in this article have been deposited in the
with a number of human pathologies, including cardiomyopathies,
Protein Data Bank, www.pdb.org (PDB ID codes 3KO0 and 3M0W).
cancer, neurodegeneration, and inflammatory disorders (1 and 6).
1To whom correspondence should be addressed. E-mail: [email protected].
For S100A4, increased protein expression correlates with a high
2Present address: Department of Biology and Biochemistry, University of Bath, Bath, BA2
incidence of metastasis and poor prognosis for a number of
7AY United Kingdom.
different cancers (7 and 8). In addition, high S100A4 expression
This article contains supporting information online at
levels contribute to fibrotic and inflammatory diseases such as
PNAS ∣ May 11, 2010 ∣ vol. 107 ∣ no. 19 ∣ 8605–8610
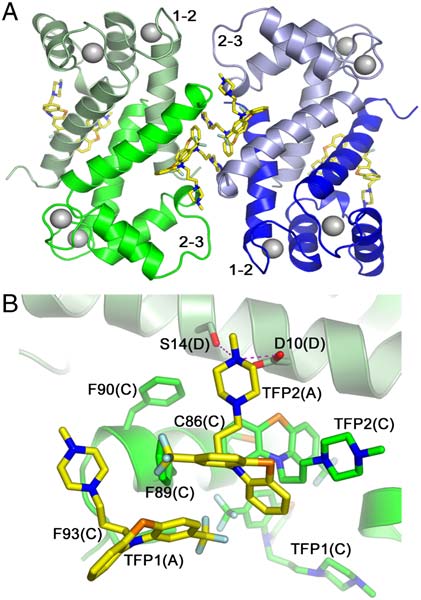

Fig. 1. Crystal packing of the Ca2þ-S100A4/TFP molecules. The protein dimers are arranged in a pentameric ring facilitated by TFP molecules (dark gray).
(A) and (B) Ribbon diagrams showing top and side views of the pentamer, respectively; (C) and (D) Corresponding space filling models. Note the large solventchannel in the interior of the pentamer.
and generous areas of the Ramachandran plot, respectively (19)
are with TFP2(A), three water molecules and the C-terminal re-
(Residues with less favorable backbone conformations
sidues of helix 4 from the symmetry-related molecule (Phe89(C),
are located in the conformationally flexible loops and the C-term-
Phe93(C), and TFP1(C)) (Fig. 2B). TFP2(A) binds with its phe-
inal tail. Most of the key structural elements in the 20 indepen-
nothiazine moiety packing against helix 4 (Ile82(A), Cys86(A),
dent chains in the asymmetric unit exhibit well defined electron
and Phe89(A)), residues in the loop between helices 2 and 3
density except for the most C-terminal residues (Asp95–Lys101)
(Leu42(A), Ser44(A), and Phe45(A)), and helix 1 of subunit B
and the loop between helices 2 and 3 (Gly47–Glu52). Further
structural description, discussion, and comparisons will be based
(Glu6(B), Leu9(B), and Asp10(B)). The methyl-piperazine ring
on the structure of the S100A4 dimer comprised of subunits A
of TFP2(A) interacts with the sidechains of the symmetry-related
and B since they display the most well defined electron density
molecules (Cys86(C), Phe89(C), TFP2(C), Asp10(D), and Ser14
(D)) and eight water molecules. There are two potential hydro-gen bonds (2.8 and 3.0 Å) between the piperazine ring atom N3
Quaternary Structure of Ca2þ-S100A4/TFP. S100 family proteins
are typically homodimers; however higher-order oligomers can
be formed under specific conditions (20–22). As described by us
previously, Ca2þ-S100A4 dimers can assemble into a continuous
superhelical arrangement due to the interaction of the C-terminal
tail with the target peptide binding cleft of symmetry-related
molecules (5). In the current crystal structure, TFP binding
results in the assembly of five Ca2þ-S100A4/TFP dimers into a
pentameric ring with a molecular point symmetry of 52 (Fig. 1).
The asymmetric unit contains two independent copies of the
pentamer. Each pentamer has inner and outer diameters of 25
and 85 Å, respectively and a thickness of 51 Å. Within each
pentamer, many of the contacts between S100A4 dimers occur
through the TFP moieties (Fig. 2A). In addition, direct contacts
are observed between residues in the loop connecting helices 1
and 2 (Gly21–Lys26), and residues from the loop connecting
helices 2 and 3 (Gly47–Glu52) in the symmetry-related molecule
(Fig. 2A). These contacts may be modest due to the significant
degree of disorder within the Gly47–Glu52 loop. Fig. 2B shows
the two TFP molecules from subunit A interacting with symme-
try-related molecule CD. The details of these interactions are
described below.
Molecular Interactions Between TFP and Ca2þ-S100A4. In the crystal,
two TFP molecules are bound per S100A4 subunit (four inhibitor
molecules per dimer), and both inhibitor molecules are well
defined in the electron density map (Fig. 3 A, B). The interactions
of the two TFP molecules (defined TFP1 and TFP2) are summar-
ized in . The phenothiazine moiety of TFP1(A) packs
against the solvent exposed side of helix 4 with the trifluoro-
methyl group pointing towards the C terminus of the helix
Fig. 2. View of the Ca2þ-S100A4/TFP dimer-to-dimer crystal interface.
(Fig. 3C). These hydrophobic contacts involve the sidechains
(A) Ribbon diagram of the AB (light and dark blue) and CD (dark and light
of Ile82(A), Met85(A), Cys86(A), and Phe89(A). The methyl-
green) S100A4 dimers. The Ca2þ atoms are shown as light gray spheres. The
piperazine ring protrudes towards the hinge region between he-
interhelical loops connecting helices 1 and 2 (1–2) and helices 2 and 3 (hinge),
lices 2 and 3, and contacts the sidechains of Ser44(A), Phe45(A),
which are involved in crystal contacts, are indicated. (B) TFP interactions withthe symmetry-related molecule. View from subunit A (light blue) towards
Leu46(A), and Gly47(A). There is a potential hydrogen bond
CD dimer. The two TFP molecules (TFP1(A) and TFP2(A)) bound to subunit
(2.9 Å) between the piperazine ring atom N3 and the carbonyl
A are shown in yellow. Helix 4 of subunit C is dark green and helix 1 of subunit
oxygen of Phe45(A). The remaining interactions of TFP1(A)
D is light green. Hydrogen bonds are shown as dashed pink lines.
Malashkevich et al.
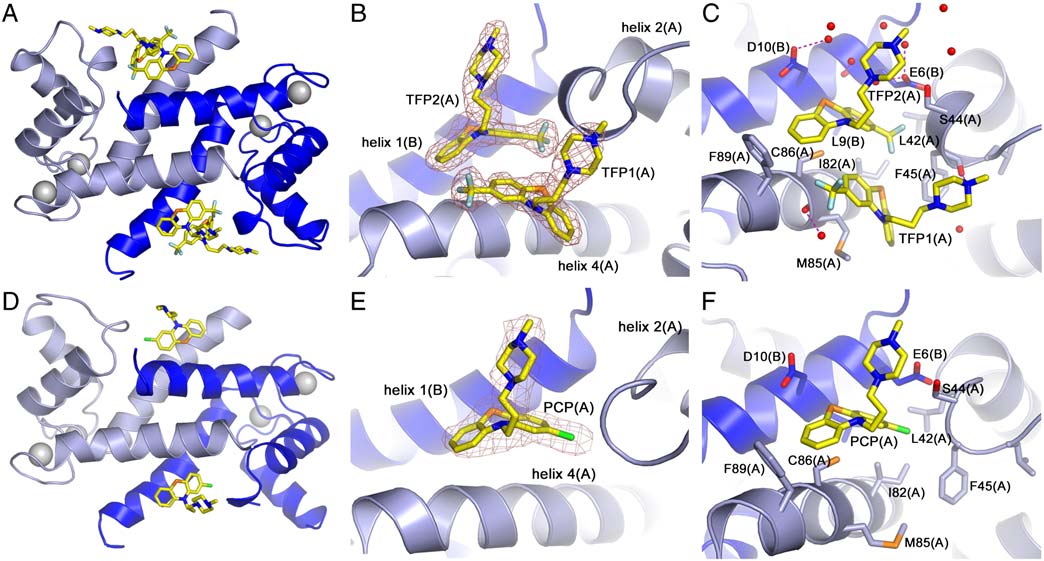

Fig. 3. Molecular interactions of Ca2þ-S100A4 and TFP or PCP. (A, D) Ribbon diagram of Ca2þ-S100A4/TFP and Ca2þ-S100A4/PCP AB dimer. Subunit A is lightblue, subunit B is dark blue, and the TFP and PCP molecules are shown in yellow. Calcium ions are shown as gray spheres. (B, E) Zoomed view of TFP and PCPmolecules in subunit A and their final refined 2Fo-Fc electron density map contoured at 1σ. (C, F). Molecular interactions of TFP and PCP molecules from subunitA. The water molecules are shown as red spheres. Hydrogen bonds are shown as dashed pink lines.
and the carboxylic group of Asp10(D), and one between the
The position and interactions of these high occu-
piperazine ring atom N2 and a water molecule (2.8 Å).
pancy PCP molecules are almost identical to the TFP2 molecules
Based on AREAIMOL (23), interactions with the native AB
and Fig. 3 D, E, F). PCP can be modeled with low oc-
dimer (including the TFP2(A) molecule) bury 58% of the solvent
cupancy in only two of the ten chains in the pentameric assembly
accessible area of TFP1(A) and 59% of the solvent accessible
at positions similar to those of TFP1.
area of TFP2(A), whereas interactions with symmetry-related
molecules (excluding waters) bury 29% and 40% of the solvent
Binding of TFP to Ca2þ-S100A4 and S100A4 Oligomerization. To deter-
accessible area, respectively ). These values indicate
mine whether the crystallographically observed interactions
that within the crystal, TFP-S100A4 interactions are strongly
between the target binding cleft of S100A4 and TFP also occur
influenced by crystal contacts, which are propagated within each
in solution, we monitored perturbations of backbone 1H-15N
pentameric ring. In total, 81% of TFP1(A) (467 Å2 out of
chemical shifts as TFP was added to S100A4. The binding was
578 Å2) and 87% of TFP2(A) (502 Å2 out of 574 Å2) solvent
Ca2þ-dependent as no chemical shift perturbations were detected
accessible areas are buried upon complex formation. Moreover,
when TFP was titrated into apo-S100A4. In the presence of Ca2þ
similar interactions are present in all the molecules in the asym-
and TFP, perturbations in 1H-15N correlations of Ca2þ-S100A4
metric unit. However, the TFP1 and TFP2 molecules assume very
were observed (>25 Hz) in the fast-exchange regime for residues
different conformations within the S100A4 binding site (
in helix 1 (Leu5, Val11, Met12, and Lys18), loop 1 (Asn30), loop 2
Superimposition of the two molecules through their phenothia-
(Leu42, Gly47, Arg49, and Asp51), helix 3 (Ala54, Phe55, Lys57,
zine moieties reveals that the tricyclic rings have opposite puckers
Leu58, and Met59), loop 3 (Glu69), and helix 4 (Leu79, Asn87,
with respect to one another. Similarly, the piperazine rings, which
and Phe89) Several of the chemical shift perturbations
are in a chair configuration, have opposing orientations.
are consistent with TFP binding to the hydrophobic target binding
cleft of S100A4 that is exposed upon Ca2þ-binding. As the TFP
Quaternary Structure of Ca2þ-S100A4/PCP. Similar to the
concentration was increased to >350 μM, the Ca2þ-S100A4 reso-
Ca2þ-S100A4/TFP structure, PCP binding results in the assembly
nances began to broaden and eventually disappeared, consistent
of five Ca2þ-S100A4/PCP dimers into a pentameric ring with a mo-
with the formation of a large complex in solution.
lecular point symmetry of 52 (). However, the asymmetric
To ascertain whether the S100A4/TFP oligomers observed by
unit of Ca2þ-S100A4/PCP contains only a single pentameric ring.
X-ray crystallography also occur in solution, we performed
Apparently, the different stoichiometry of PCP binding and crys-
analytical sedimentation studies. Titration of Ca2þ-S100A4 with
tallization conditions produced a unit cell with only half the volume
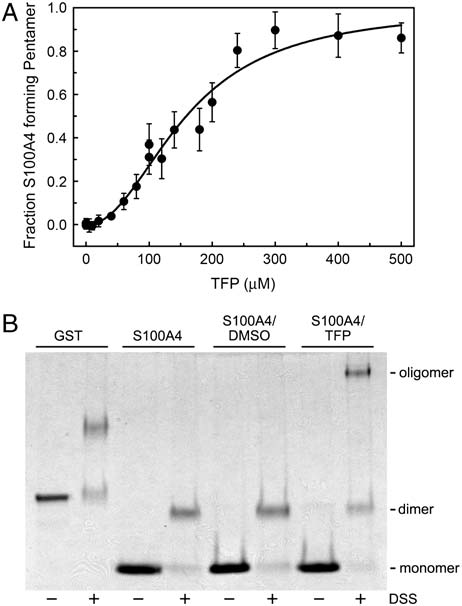
TFP resulted in the formation of a complex with a weight-average
of that observed for the Ca2þ-S100A4/TFP crystals. Otherwise,
molecular weight of 133; 107 " 8; 671 Da (Fig. 4A). An S100A4
the structures of the Ca2þ-S100A4/TFP and Ca2þ-S100A4/PCP
oligomer comprised of five dimers and 20 TFP molecules has a
structures are similar with a rms deviation of 0.42 Å and 0.58 Å
calculated molecular weight of 124,123 Da, which is in good
for all Cα atoms for monomers and pentameric rings, respectively.
agreement with the experimentally determined molecular weight
of the Ca2þ-S100A4/TFP oligomers. These observations suggest
Molecular Interactions Between PCP and Ca2þ-S100A4. In contrast
that under the conditions of our sedimentation studies, TFP
to TFP, only a single high occupancy PCP molecule is present
induces a S100A4 oligomer similar to that observed by crystallo-
in each S100A4 subunit (two inhibitor molecules per dimer)
graphy. Moreover, an examination of the Hill coefficient
Malashkevich et al.
PNAS ∣ May 11, 2010 ∣ vol. 107 ∣ no. 19 ∣ 8607
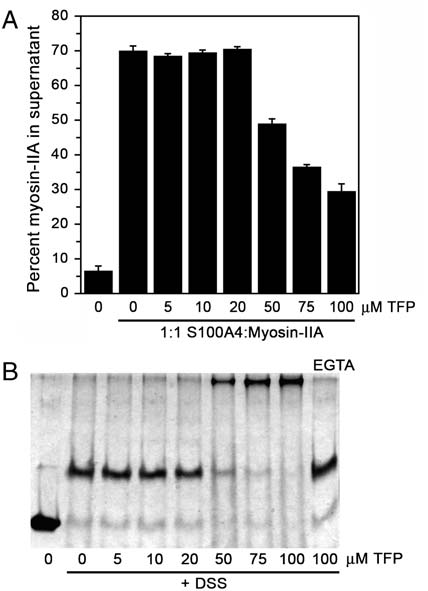

rods polymerize into filaments, and Ca2þ-dependent binding of
S100A4 to myosin-IIA almost completely disassembles these
preformed filaments (18 and 24). We also reported that 100 μM
TFP completely blocks the ability of S100A4 to depolymerize myo-
sin-IIA filaments (18). To test whether lower TFP concentrations
were capable of inhibiting S100A4-mediated depolymerization of
myosin-IIA filaments, we assayed a range of TFP concentrations in
the promotion of disassembly assay. Concentrations of TFP up to
20 μM had no effect on S100A4's myosin-IIA depolymerizing
activity (Fig. 5A). Only at TFP concentrations of ≥50 μM was sig-
nificant inhibition of S100A4 activity detected. Notably, parallel
cross-linking studies showed that at TFP concentrations that inhi-
bit S100A4 activity, there is significant S100A4 oligomerization
into high molecular weight complexes (Fig. 5B). In addition,
the formation of large S100A4 oligomers by TFP requires
Ca2þ-binding, as only S100A4 dimers were detected in the absence
In our original screen against a library of FDA-approved
drugs, we identified six phenothiazines as inhibitors of myosin-
IIA associated S100A4 function. To determine if the TFP-induced
oligomerization of S100A4 is specific to this phenothiazine, or is
a general feature of this class of compounds, we performed che-
mical cross-linking assays under the same conditions utilized for
TFP. All the phenothiazines tested, which include, flupenthixol,
fluphenazine, chlorprothixene, prochlorperazine, and perphena-
zine induced S100A4 oligomerization, albeit to varying extents
(). Oligomeric S100A4 intermediates of 31, 46, and
66 kDa were observed for all of the phenothiazines. However, only
chlorprothixene, and to a lesser extent prochlorperazine, induced
the formation of large S100A4 oligomers (
Fig. 4. TFP induces S100A4 oligomerization. (A) Plot of fraction S100A4
∼120 kDa) as seen with
pentamer versus TFP concentration. Sedimentation equilibrium data were
TFP. The observation that 100 μM PCP does not promote the same
collected at 25 °C at a concentration of 80 μM S100A4 subunit. (B) Chemical
cross-linking of Ca2þ-S100A4. Coomassie-stained SDS-PAGE of GST control,S100A4 alone, S100A4 þ DMSO, and S100A4 þ TFP. Monomeric and dimeric
S100A4 have apparent molecular weights of approximately 11.5 kDa and23 kDa, respectively.
(nH ¼ 2.4 " 0.3) indicates that S100A4 oligomerization is coop-
erative with a midpoint of 150 μM TFP.
Since the sedimentation equilibrium experiments required
significantly higher S100A4 subunit concentrations than those
used to evaluate S100A4 activity in our biochemical assays, we
used chemical cross-linking to examine whether TFP promotes
S100A4 oligomerization under conditions in which S100A4
depolymerizes myosin-IIA filaments. In the presence of the che-
mical cross-linker disuccinimidylsuberate (DSS) (but no TFP), a
prominent S100A4 band was detected at approximately 23 kDa,
consistent with a dimer (Fig. 4B). In the presence of 100 μM TFP,
a band of approximately 120 kDa was observed in the DSS-
treated sample, which is consistent with the oligomer detected
in sedimentation and X-ray studies. Larger oligomers were not
detected by SDS-PAGE (suggesting the formation
of a distinctly sized S100A4/TFP complex.
Sedimentation equilibrium analysis of this crosslinked species
revealed a stable, homogenous S100A4 oligomer with a weight-
average molecular weight of 143; 372 " 7; 462 Da. DSS is a
homobifunctional cross-linker that contains an amine-reactive
N-hydroxysuccinimide ester at each end of an 8-carbon spacer
arm. Typically the N terminus of the polypeptide chain and
the lysine sidechain are the targets of DSS cross-linking. Each
S100A4 subunit contains twelve lysine residues, all of which
are solvent accessible. Given the uncertainty of the number of
Fig. 5. S100A4-mediated myosin-IIA depolymerization in the presence of
cross-links in the S100A4-TFP-DSS complex, the experimental
TFP. (A) Quantification of the myosin-IIA disassembly assay. In the absence
molecular weight is in good agreement with the calculated mo-
of S100A4, 7% of the myosin-IIA rods are recovered in the supernatant;
lecular weight of an oligomer comprised of five S100A4 dimers,
whereas in the presence of S100A4, 70% of the myosin-IIA rods are presentin the supernatant. Values represent the mean
20 TFP molecules, and multiple cross-links.
" sd from two independent
experiments. (B) Chemical cross-linking of Ca2þ-S100A4 in the presence of
Our previous studies showed that under the conditions of our
increasing concentrations of TFP (0–100 μM). The last lane shows the products
sedimentation assay approximately 75–80% of the myosin-IIA
of the cross-linking reaction in the presence of EGTA and 100 μM TFP.
Malashkevich et al.
extent of oligomerization as TFP is consistent with our previous
to the shorter loop between helices 2 and 3 and the lack of a
report that, 100 μM PCP inhibits S100A4's depolymerizing activity
second subunit (troponin C is a monomer) (Fig. 6A). Interest-
to a lesser extent than 100 μM TFP in the myosin-IIA disassembly
ingly, in the crystal structure of the troponin C/TFP complex, di-
assay (18), and, as evidenced by the S100A4/PCP structure,
merization occurs via interactions between the two pairs of TFP
suggests that higher PCP concentrations are required to induce
molecules, which "glue" the two troponin C monomers together.
the formation of the pentameric S100A4 species.
However, the disposition of the two troponin C subunits is quite
different from that observed in a typical S100 family dimer. In the
1∶2 Ca2þ-calmodulin/TFP complex (Fig. 6B), TFP1 binds deep in
The current study represents one of the few examples of a
the cleft formed by the C-terminal EF-hand similar to troponin C,
S100-small molecule inhibitor structure, and provides a detailed
whereas TFP2 occupies an interdomain site. One consequence of
description of a phenothiazine binding to an S100 protein. Previous
TFP binding is to bring the N- and C-terminal domains of calmo-
studies reported on the Ca2þ-dependent interaction between
dulin together so that the protein assumes a compact globular
S100A1/S100A1 (S100a), S100A1/S100B (S100a ), and S100B/
conformation similar to that observed in structures of Ca2þ-cal-
S100B (S100b) with TFP (25 and 26) as well as other phenothia-
modulin/peptide complexes (31 and 32). The compact structure
zines (27), but did not delineate the binding site or residues
of calmodulin prevents the two bound TFP molecules from
involved in binding. Our studies demonstrate that each S100A4
clustering with TFP molecules from the symmetry-related
subunit binds two TFP molecules in the target binding cleft formed
protein chains.
by the hinge and helices 3 and 4.
At present, S100A4 is the only EF-hand containing protein in
In addition to the S100 proteins, other EF-hand containing
which phenothiazines (TFP, PCP) induce the formation of higher-
proteins bind TFP. For example, troponin C binds two TFP
order oligomers. Both sedimentation equilibrium and chemical
molecules (PDB 1WRK) and calmodulin binds TFP with a range
cross-linking studies demonstrate that Ca2þ-S100A4/TFP com-
of stoichiometries; 1∶1 (PDB 1CTR), 1∶2 (PDB 1A29), and 1∶4
plexes can form oligomers comprised of at least five S100A4 di-
(PDB 1LIN) (28–30). Even though S100A4, troponin C, and cal-
mers in solution. We previously reported that TFP binds to the
modulin are built upon the same basic four-helical structural
Mero-S100A4 with an EC50 value of 55 " 2.6 μM (18). Notably,
module, the architectures of the TFP binding pockets, and the
our cross-linking experiments revealed that TFP concentrations
positions and orientations of the bound TFP molecules are quite
of 50 μM are sufficient to induce S100A4 oligomerization and at
different amongst the three proteins (Fig. 6). For example, in
this TFP concentration we first observe inhibition of S100A4's
the Ca2þ-troponin C/TFP complex, the two TFP molecules are
myosin-IIA-associated activities. Based on these findings, we
positioned deeper in the cleft formed by helices 3 and 4 due
propose that rather than directly competing with myosin-IIA,
our structural, biophysical, and biochemical data support a model
in which phenothiazines disrupt the S100A4/myosin-IIA interac-
tion by sequestering S100A4 into a large well defined oligomer. A
comparison of the residues that exhibit chemical shift perturba-
tions following titration with TFP or the MIIA1908–1923 peptide (5)
indicates that the two ligands occupy overlapping, but noniden-
tical sites within the hydrophobic target binding cleft. (Table 1,
While these observations might be consistent with
the direct competition of TFP with the myosin-IIA peptide for
S100A4 binding, it is important to note that disassembly assays
use the physiologically relevant dimeric myosin-IIA coiled-coil
that is also likely to be bivalent. Due to enhanced contact surface
and avidity, the full-length myosin-IIA tail may not be easily
displaced by TFP binding to S100A4.
An examination of available S100 protein/target peptide struc-
tures reveals that targets can bind in a variety of orientations and
conformations (). A hypothetical model of the S100A4/
myosin-IIA pentamer suggests that inhibitor-induced oligomeri-
zation may preclude efficient recognition of the myosin-IIA
coiled-coil due to unfavorable steric interactions. Myosin-IIA
heavy chains bound to neighboring S100A4 subunits would cross
inside the pentameric ring resulting in considerable steric clash
In addition, the interior of the ring would have to
accommodate an additional 35 residues from the C-terminal tail-
piece of each myosin-IIA heavy chain (total 350 residues). There-
fore, the totality of our data supports a more complex model in
Table 1. Comparison of chemical shift perturbations followingaddition of TFP or the MIIA1908–1923 peptide to Ca2þ-S100A4 *.
Secondary structure
L5, V11, M12, L18
V11, M12, S14, F16
S20, F27, K28, N30
Fig. 6. Structural alignment of protein/TFP complexes. (A) Overlay of the
L42, G47, R49, D51
E41, G47, K48, T50, D51
Ca2þ-S100A4/TFP and troponin C/TFP complexes (PDB 1WRK). Subunit A of
A54, F55, K57, L58, M59
S100A4 is light blue and the TFP molecules are shown in yellow. Troponin
C is red and the two bound TFP molecules are light red. Calcium atoms
F78, L79, M85, C86
are presented as corresponding colored spheres. (B) Overlay of the
Ca2þ-S100A4/TFP and calmodulin/TFP 1∶2 complexes (PDB 1A29). Calmodulinand the two bound TFP molecules are colored green.
*Shared residues are in bold italics.
Malashkevich et al.
PNAS ∣ May 11, 2010 ∣ vol. 107 ∣ no. 19 ∣ 8609
which TFP may initially compete with myosin-IIA for the same
Materials and Methods
binding pocket, but TFP-driven oligomerization prevents S100A4
Structure Determination. Recombinant human S100A4 was purified as de-
binding to myosin-IIA. Additionally, the S100A4/PCP structure
scribed previously (24). Crystals of S100A4 bound to TFP or PCP were obtained
and the similar cross-linking behavior induced by a wide range
by sitting drop vapor diffusion at 293 K. All structures were solved by mole-cular replacement and refined by standard methods, resulting in R
of phenothiazines suggest that oligomerization is an important
values of 20.6%/25.9% and 25.2%/30.2% for the S100A4/TFP and S100A4/PCP
mechanistic feature of phenothiazine-induced S100A4 inhibition.
complexes, respectively (). Details of the structure determination are
A number of small molecules have been described that inhibit
protein function by altering the oligomerization equilibrium ofthe target protein. For example, a small molecule inhibitor of
NMR Spectroscopy. 1H and 15N resonances Ca2þ-S100A4 were followed during
titrations with TFP at 37 °C using two-dimensional 1H-15N heteronuclear sin-
α promotes subunit dissociation from the biologically active
gle quantum correlation (HSQC) spectra, and their assignments were con-
trimer to stabilize an inactive dimer (33). For porphobilinogen
firmed with a three-dimensional 15N-edited NOESY-HSQC experiment.
synthase, binding of the small molecule morphlock-1, shifts the
Proton chemical shifts were reported with respect to the H2O or HDO signal
oligomeric equilibrium from the active octamer to a low activity
taken as 4.658 ppm relative to external trimethylsilyl-2,2,3,3-tetradeutero-
hexamer (34). In the case of HIV-1 integrase, inhibitory peptides
propionic acid (TSP) (0.0 ppm), and the 15N chemical shifts were indirectlyreferenced as described previously using the following ratio of the zero-point
shift the oligomerization equilibrium from the active dimer to an
frequency: 0.10132905 for 15N to 1H. Details are provided in .
inactive tetramer (35). Although the inactive tetramer has notbeen characterized structurally, the peptide is thought to induce
Biochemical Assays. Promotion of disassembly assays were performed as
the formation of a nonnatural tetramer (35). The recent charac-
described previously (24). S100A4 cross-linking experiments with performed
terization of S100B, S100A8/A9 and S100A12 as well-defined
at 25 °C with 5 mM disuccinimidylsuberate and 100–500 μM phenothiazines.
Details are provided in
oligomers comprised of two to four S100 dimers (20–22), suggests
that the propensity to form higher-order structures may be a
Analytical Ultracentrifugation. Sedimentation equilibrium experiments were
common feature of S100 proteins. This characteristic is also
performed at 25 °C with a Beckman XL-I analytical ultracentrifuge using
shared by S100A4 as we and others demonstrated that S100A4
the absorbance optics and Ti60 rotor. Details are provided in the .
can form tetramers or higher-order oligomers in the absenceof added compounds (5 and 36). We propose that TFP-mediated
ACKNOWLEDGMENTS. We acknowledge the staff of the LRL Collaborative
Access Team beamline at the Advanced Photon Source and the X29 beamline
oligomerization of S100A4 is another example of stabilization of
at the National Synchrotron Light Source. This work was supported with
an inactive, nonnatural oligomer. The formation of inactive
National Institutes of Health Grants CA129598 (to A.R.B.), and GM58888
S100A4 oligomeric assemblies may be a useful and unique strat-
and CA107331 (to D.J.W.). We acknowledge support from the Albert Einstein
College of Medicine Cancer Center (National Cancer Institute (NCI) Grant
egy for inhibitor development.
1. Marenholz I, Heizmann CW, Fritz G (2004) S100 proteins in mouse and man: from
19. Laskowski R, MacArthur M, Moss D, Thornton J (1993) PROCHECK: a program to check
evolution to function and pathology (including an update of the nomenclature).
the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291.
Biochem Biophys Res Commun 322:1111–1122.
20. Ostendorp T, et al. (2007) Structural and functional insights into RAGE activation by
2. Zimmer DB, Cornwall EH, Landar A, Song W (1995) The S100 protein family: history,
multimeric S100B. Embo J 26:3868–3878.
function, and expression. Brain Res Bull 37:417–429.
21. Korndorfer IP, Brueckner F, Skerra A (2007) The crystal structure of the human (S100A8/
3. Drohat AC, Baldisseri DM, Rustandi RR, Weber DJ (1998) Solution structure of
S100A9)2 heterotetramer, calprotectin, illustrates how conformational changes of
calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance
interacting alpha-helices can determine specific association of two EF-hand proteins.
spectroscopy. Biochemistry 37:2729–2740.
J Mol Biol 370:887–898.
4. Sastry M, et al. (1998) The three-dimensional structure of Ca(2+)-bound calcyclin:
22. Moroz OV, Blagova EV, Wilkinson AJ, Wilson KS, Bronstein IB (2009) The crystal
implications for Ca(2+)-signal transduction by S100 proteins. Structure 6:223–231.
structures of human S100A12 in apo form and in complex with zinc: new insights into
5. Malashkevich VN, et al. (2008) Structure of Ca2+-bound S100A4 and its interaction
S100A12 oligomerisation. J Mol Biol 391:536–551.
with peptides derived from nonmuscle myosin-IIA. Biochemistry 47:5111–5126.
23. Collaborative Computational Project N (1994) The CCP4 suite: programs for protein
6. Heizmann CW, Ackermann GE, Galichet A (2007) Pathologies involving the S100
crystallography. Acta Crystallogr D 50:760–763.
proteins and RAGE. Subcell Biochem 45:93–138.
24. Li ZH, Spektor A, Varlamova O, Bresnick AR (2003) Mts1 regulates the assembly of
7. Helfman DM, Kim EJ, Lukanidin E, Grigorian M (2005) The metastasis associated pro-
nonmuscle myosin-IIA. Biochemistry 42:14258–14266.
tein S100A4: role in tumour progression and metastasis. Br J Cancer 92:1955–1958.
25. Pingerelli PL, Mizukami H, Mooney MJ, Schlaepfer AL (1989) Spectral studies of the
8. Garrett SC, Varney KM, Weber DJ, Bresnick AR (2006) S100A4, a mediator of metas-
Ca2+-dependent interaction of trifluoperazine with S100b. J Protein Chem 8:183–196.
tasis. J Biol Chem 281:677–680.
26. Pingerelli PL, Mizukami H, Wagner AS, Bartnicki DE, Oliver JP (1990) Investigation of
9. Schneider M, Hansen JL, Sheikh SP (2008) S100A4: a common mediator of epithelial-
the Ca2(+)-dependent interaction of trifluoperazine with S100a: a 19F NMR and
mesenchymal transition, fibrosis and regeneration in diseases?. J Mol Med 86:507–522.
circular dichroism study. J Protein Chem 9:169–175.
10. Grigorian M, Ambartsumian N, Lukanidin E (2008) Metastasis-inducing S100A4
27. Marshak DR, Watterson DM, Van Eldik LJ (1981) Calcium-dependent interaction of
protein: implication in non-malignant human pathologies. Curr Mol Med 8:492–496.
S100b, troponin C, and calmodulin with an immobilized phenothiazine. Proc Natl
11. Kriajevska MV, et al. (1994) Non-muscle myosin heavy chain as a possible target for
Acad Sci USA 78:6793–6797.
protein encoded by metastasis-related mts-1 gene. J Biol Chem 269:19679–19682.
12. Takenaga K, et al. (1994) Binding of pEL98 protein, an S100-related calcium-binding
28. Vertessy BG, et al. (1998) Simultaneous binding of drugs with different chemical struc-
protein, to nonmuscle tropomyosin. J Cell Biol 124:757–768.
tures to Ca2+-calmodulin: crystallographic and spectroscopic studies. Biochemistry
13. Watanabe Y, et al. (1993) Calvasculin, as a factor affecting the microfilament
assemblies in rat fibroblasts transfected by src gene. FEBS Lett 324:51–55.
29. Cook WJ, Walter LJ, Walter MR (1994) Drug binding by calmodulin: crystal structure of
14. Kriajevska M, et al. (2002) Liprin beta 1, a member of the family of LAR
a calmodulin-trifluoperazine complex. Biochemistry 33:15259–15265.
transmembrane tyrosine phosphatase-interacting proteins, is a new target for the
30. Vandonselaar M, Hickie RA, Quail JW, Delbaere LT (1994) Trifluoperazine-induced
metastasis-associated protein S100A4 (Mts1). J Biol Chem 277:5229–5235.
conformational change in Ca(2+)-calmodulin. Nat Struct Biol 1:795–801.
15. Grigorian M, et al. (2001) Tumor suppressor p53 protein is a new target for the me-
31. Ikura M, et al. (1992) Solution structure of a calmodulin-target peptide complex by
tastasis-associated Mts1/S100A4 protein: functional consequences of their interaction.
multidimensional NMR. Science 256:632–638.
J Biol Chem 276:22699–22708.
32. Meador WE, Means AR, Quiocho FA (1993) Modulation of calmodulin plasticity in
16. Semov A, et al. (2005) Metastasis-associated protein S100A4 induces angiogenesis
molecular recognition on the basis of X-ray structures. Science 262:1718–1721.
through interaction with Annexin II and accelerated plasmin formation. J Biol Chem
33. He MM, et al. (2005) Small-molecule inhibition of TNF-alpha. Science 310:1022–1025.
34. Lawrence SH, et al. (2008) Shape shifting leads to small-molecule allosteric drug
17. Dukhanina EA, et al. (2009) Opposite roles of metastasin (S100A4) in two potentially
discovery. Chem Biol 15:586–596.
tumoricidal mechanisms involving human lymphocyte protein Tag7 and Hsp70. Proc
35. Hayouka Z, et al. (2007) Inhibiting HIV-1 integrase by shifting its oligomerization
Natl Acad Sci USA 106:13963–13967.
equilibrium. Proc Natl Acad Sci USA 104:8316–8321.
18. Garrett SC, et al. (2008) A biosensor of S100A4 metastasis factor activation: inhibitor
36. Gingras AR, et al. (2008) Crystal structure of the Ca(2+)-form and Ca(2+)-binding
screening and cellular activation dynamics. Biochemistry 47:986–996.
kinetics of metastasis-associated protein, S100A4. FEBS Lett 582:1651–1656.
Malashkevich et al.
Source: https://www.ibbr.umd.edu/sites/default/files/event/Qiong%2520Zhang%252010-19-12.pdf
VOL. 1 N0 3. JAN -JUNE, 2004 An interview with the D.G -Nigerian Institute of Me dical Research By NIMRNEWS Editorial Board NIMRNEWS: Sir, what will you death. Talking about consider as your achievement in the past subvention, for the This edition of NIMRNews focused more on facts finding mission by engaging the DIRECTOR-GENERAL: The first thing
Ausgabe 43 Erntedank — 1. Advent 2010 Gemeindebrief der Scheidegg - Weiler wir wollen helfen in aller Not, wir wollen schaffen, die Kraft gibst Du, wir wollen lieben, Herr hilf dazu. Amen. Umsichtig lenkte der Fahrer durch Vor einiger Zeit ging für mich ein die engen Kurven und erklärte mir Kindheitstraum in Erfüllung. Als nebenbei die Technik. Stolz und Erstklässler habe ich mir vom glücklich stieg ich unten aus. Taschengeld jede Woche kleine Wie geht es uns mit unseren Spielzeugautos gekauft, und beson-












