
Cialis ist bekannt für seine lange Wirkdauer von bis zu 36 Stunden. Dadurch unterscheidet es sich deutlich von Viagra. Viele Schweizer vergleichen daher Preise und schauen nach Angeboten unter dem Begriff cialis generika schweiz, da Generika erschwinglicher sind.
Doi:10.1530/rep-14-0126
Maternal age effect on mouse oocytes: new biologicalinsight from proteomic analysis
Caroline Schwarzer*, Marcin Siatkowski1,2,*, Martin J Pfeiffer, Nicole Baeumer3,Hannes C A Drexler4, Bingyuan Wang, Georg Fuellen1,2 and Michele Boiani
Max Planck Institute for Molecular Biomedicine, Ro¨ntgenstraße 20, D-48149 Mu¨nster, Germany,
1DZNE, German Centre for Neurodegenerative Disorders, Gehlsheimer Straße 20, D-18147 Rostock, Germany,
2Institute for Biostatistics and Informatics in Medicine and Ageing Research, Rostock University Medical Center,Ernst Heydemann-Str. 8, D-18057 Rostock, Germany, 3Arrows Biomedical GmbH, Gievenbecker Weg 11,D-48149 Mu¨nster, Germany and 4Max Planck Institute for Molecular Biomedicine, Bioanalytical Mass SpectrometryFacility, Ro¨ntgenstraße 20, D-48149 Mu¨nster, Germany
Correspondence should be addressed to M Boiani; Email:
[email protected] or to
G Fuellen; Email:
[email protected]
*(C Schwarzer and M Siatkowski contributed equally to this work)
The long-standing view of ‘immortal germline vs mortal soma' poses a fundamental question in biology concerning how oocytes age inmolecular terms. A mainstream hypothesis is that maternal ageing of oocytes has its roots in gene transcription. Investigating the proteinsresulting from mRNA translation would reveal how far the levels of functionally available proteins correlate with mRNAs and would offernovel insights into the changes oocytes undergo during maternal ageing. Gene ontology (GO) semantic analysis revealed a high similarityof the detected proteome (2324 proteins) to the transcriptome (22 334 mRNAs), although not all proteins had a cognate mRNA.
Concerning their dynamics, fourfold changes of abundance were more frequent in the proteome (3%) than the transcriptome (0.05%),with no correlation. Whereas proteins associated with the nucleus (e.g. structural maintenance of chromosomes and spindle-assemblycheckpoints) were largely represented among those that change in oocytes during maternal ageing; proteins associated with oxidativestress/damage (e.g. superoxide dismutase) were infrequent. These quantitative alterations are either impoverishing or enriching.
Using GO analysis, these alterations do not relate in any simple way to the classic signature of ageing known from somatic tissues.
Given the lack of correlation, we conclude that proteome analysis of mouse oocytes may not be surrogated with transcriptome analysis.
Furthermore, we conclude that the classic features of ageing may not be transposed from somatic tissues to oocytes in a one-to-onefashion. Overall, there is more to the maternal ageing of oocytes than mere cellular deterioration exemplified by the notoriousincrease of meiotic aneuploidy.
Reproduction (2014) 148 55–72
(Thus, ageing of oocytes may
The ‘maternal age effect' in reproduction, characterized
be viewed as a life-long maintenance of cellular
by a negative relationship between maternal age and
homeostasis in the same cell, unlike ageing of the male
reproductive success, is a poorly understood phenom-
germline. Some molecular factor(s) within oocytes might
enon. Several hypotheses have been proposed. While
deteriorate as the potential mother ages, compromising,
the receptivity of the uterus and the ovarian reserve of
for example, the function of the chromosomal apparatus
follicles can explain the ‘maternal age effect' in part, it is
or the ability to scavenge reactive oxygen species
known that age-related decline of a female's fertility is
resulting from mitochondrial reactions (
also rooted in the quality and developmental potential
of her oocytes. Unlike the male germline, the bulk of
The synthesis of the aforementioned molecular
oocytes do not have a gonial stem cell population.
factor(s) relies on gene transcription that oocytes largely
Oocytes spend most of their time quiescent in primordial
perform during follicular growth prior to ovulation, and
follicles, mature over days or weeks during follicular
the transcriptional activities of oocytes may be influ-
growth and then become quiescent again near the
enced by maternal age. Microarray and RT-PCR methods
time of ovulation, when gene transcription is silenced
have revealed that maternal ageing is accompanied
q 2014 Society for Reproduction and Fertility
ISSN 1470–1626 (paper) 1741–7899 (online)
Online version via www.reproduction-online.org
C Schwarzer, M Siatkowski and others
by changes in the levels of oocytic mRNAs involved in
and should harbour the majority of all oocyte proteins
mitochondrial function, apoptosis, oxidative stress, cell
(although with different relative abundances). Practically,
cycle regulation, chromosome stability and epigenetic
F9 cells allow for the efficient metabolic labelling of the
modification, in both mouse and human oocytes
SILAC reference in vitro, overcoming the difficulty of
directly labelling oocytes in vivo.
We analysed maternal ageing of mouse oocytes on the
protein level, using SILAC technology and high-
However, searching these studies for culprits
resolution MS, to define its signature at a level closer
elicits a list of candidate genes that is quite short,
to phenotype than mRNA. These oocytes had aged up to
featuring Bcl2 and Bax (mitochondrial function and
1 year inside the ovaries and were analysed immediately
apoptosis); Txndc9 (Apacd), Sod1 and Txn1 (oxidative
after ovulation so as to appreciate the effect of maternal
stress); Mad2l1 (Mad2) and Bub1 (spindle assembly
age on the oocyte. This should not be confused with the
checkpoint (SAC)); Atrx, Brca1, Numa1 and Smc1b
post-ovulatory ageing of oocytes in the oviduct or in a
(spindle assembly and chromosome integrity/stability);
culture medium (We compared
and Dmap1, Dnmt1, Dnmt3A and Hdac1/2 (epigenetic
and confronted the quantitative protein data with the
modification). Furthermore, the fold-change of these
predictions of mRNA ageing studies of oocytes and
mRNAs in oocytes during maternal ageing is low
somatic cells. The analysis of two maternal age
(sometimes as low as 1.4- to 1.5-fold).
transitions (puberty to mature age, first; mature age to
The hypothesis that maternal ageing of oocytes has its
climacterium, second) allowed us to disregard proteins
roots in aberrant gene transcription, as opposed to other
that keep steady throughout life and to focus on those
steps of the gene expression cascade, is largely
proteins whose abundance is changing, i.e. age
unproven and is difficult to reconcile with the transcrip-
regulated in the second age transition. Overall, corre-
tional silencing that occurs in oocytes near the time of
lation between quantitative changes in the proteome and
ovulation. Advances in ‘omics' research show that
in the transcriptome was nearly nil. Gene expression and
intermediate steps of the gene expression cascade,
gene ontology (GO) analyses revealed a distinction
as well as post-translational protein modification and
in the cellular components and biological processes
degradation, can affect levels of functionally available
(BPs) affected by ageing in oocytes compared to somatic
proteins independent of transcription (
cells. Proteins associated with the nucleus were featured
Thus, we consider that the protein level may offer
predominantly among those that changed (declined) in
novel insights into the changes in gene expression that
oocytes during maternal ageing, thereby fulfilling a
oocytes undergo during maternal ageing. The proteome,
classic expectation of oocyte ageing, i.e. progressive loss
an accessible ‘missing link' between transcriptome and
of precision in chromosome maintenance and segre-
phenotype, can be analysed using high-resolution mass
gation. By contrast, the number of changing proteins
spectrometry (MS)-based proteomics, which allows
associated with oxidative stress/damage was very small.
for the accurate identification of thousands of proteins
Our first conclusion is that proteome analysis of mouse
in somatic tissues during in vivo ageing (
oocytes may not be surrogated with transcriptome
analysis: the two molecular portraits do not correlate.
Previous MS studies on mice analysed the proteome of
Our second conclusion is that the classic features of
mature oocytes and
ageing may not be transposed from somatic tissues
processes of oocyte maturation (
to oocytes in a one-to-one manner. Overall, there is more
Monti et al., 2013) in
to the maternal ageing of oocytes than a mere cellular
young donors, but not oocyte quality and composition
deterioration exemplified by the notorious increase in
during maternal ageing. As MS is not inherently quan-
meiotic aneuploidy. This is in accord with our functional
titative, relied on the simultaneous
observations of the increased ability of old oocytes to
comparison of signal intensities between replica samples
support blastocyst formation, although reduced post-
from young donors, in a so-called label-free approach that
implantation development is predicted as a result of
requires high numbers of oocytes. Scarce samples, such as
oocytes from aged donors, are hardly amenable to thereplica collection of hundreds if not thousands of oocytes.
These scarce specimens can be quantified using a defined
Materials and methods
amount of isotopically labelled reference, which is added(‘spike-in') to the non-labelled oocyte lysate prior to
Gene, mRNA and protein nomenclature
processing for MS. This method is called SILAC (stable
We followed the rules of the Jackson Laboratory published as of
isotope labelling of amino acids in cell culture;
). F9 embryonal carcinoma (EC) cells are
Briefly, gene names are written
appropriate as a labelled reference for oocytes as they can
in italic format (e.g. Hprt), mRNA names are written in italic
easily be cultured feeder-free, have stem cell properties
format and also specified to be mRNA (e.g. Hprt mRNA),
Reproduction (2014) 148 55–72
SILAC proteome analysis of mouse oocyte ageing
protein names are not written in italic format and only
uppercase letters were used (e.g. HPRT).
For each age group, 20 oocytes were collected in biologicaltriplicates. Microarray data of the MII oocytes were obtained
using the Agilent platform, as described )and deposited at
This mouse ageing study was performed in accordance with
the recommendations of the Federation of Laboratory Animal
RNA was extracted using the ZR RNA Microprep Kit (Zymo
Science Associations (FELASA) and with the ethical permit
Research Corporation, Irvine, CA, USA). A two-round linear
issued by the Landesamt fu¨r Natur, Umwelt und Verbrau-
amplification protocol employing a linear two-step TargetAmp
cherschutz (LANUV) of the state of North Rhine-Westphalia,
2-Round Biotin-aRNA Amplification Kit 3.0 (Epicentre, Madi-
Germany (permit number: 87-51.04.2010.A160). Every effort
son, Wisconsin) was used to generate biotin-labelled cRNA.
was made to preserve animal welfare during the prolonged
Similar to other studies, cDNA was synthesized using mRNA-
period of time necessary to complete the study.
specific poly(dT) primers binding the poly(A) tail of mRNA). It is possible that
Oocyte collection
amplification with poly(dT) primers distorts the representationof the original mRNA population by amplifying preferentially
Germinal vesicle (GV) B6C3F1 oocytes were collected from the
those mRNAs that have a longer poly(A) tail instead of those
ovarian follicles 48 h after injection of pregnant mare serum
mRNAs with short or no poly(A) tail
gonadotrophin (Intergonan, Intervet, Unterschleißheim,
However, the use of poly(dT) primers ensures that only those
Germany). Metaphase II (MII) oocytes were collected from the
transcripts are amplified that are amenable to translation at the
oviductal ampullae 14 h after the injection of human chorionic
time of sampling.
gonadotrophin (Ovogest, Intervet), as described (Oocytes used were ovulated from 3 week (pubertal), 8G1week (mature) and 58
Isotopic labelling
G10 week (climacteric) old B6C3F1 mice.
F9 EC cells were grown for several passages in RPMI 1640
Karyotype analysis
medium (PAA, Co¨lbe, Germany), supplemented with 10%dialysed FCS (Sigma), heavy amino acids 13C15
6 N2-L-lysine (K8)
MII oocytes were processed for chromosome counting by
6 N4-L-arginine (R10; Silantes, Martinsried, Germany)
air-drying and Hoechst dye staining, as described
and glutamine and the antibiotics penicillin and streptomycin
. As chromosome spreads with fewer than 20 chromosomes
(Gibco, Life Technologies, Darmstadt, Germany). Labelling
may be the result of chromosome loss during the spreading
efficiency was examined by small-scale in-solution digests of
procedure, only supernumerary chromosomes are considered
the heavy labelled F9 cells and analysed by LC-MS/MS (see
reliable. Therefore, the degree of aneuploidy is measured by the
below). Labelled EC cells were used as a spike-in standard
ratio of chromosome spreads with O20 chromosomes over
when the 100 most intense proteins showed a labelling
chromosome spreads with R20 chromosomes.
efficiency O97.8% (see see sectionon given at the end of this article). In orderto quantify the labelling efficiency, light F9 cells were grown in
Embryo production
the same medium except for the heavy amino acids, which
Embryos were produced by intracytoplasmic sperm injection
were replaced by the conventional amino acids 12C14
(ICSI), somatic cell nuclear transfer (SCNT) or parthenogenesis
lysine (K0) and 12C14
6 N4-L-arginine (R0; Sigma), labelled as
(PA) of MII B6C3F1 oocytes and cultured in a-MEM medium, as
‘light' to discriminate them from their ‘heavy' counterparts.
described Nucleus donors were C57Bl/6Jfor ICSI and OG2 (B6;CBA-Tg(Pou5f1-EGFP)2Mnn/J) for SCNT.
Protein isolation, fractionation and MS
Zona-free oocytes and F9 cells were lysed in SDS lysis buffer
Confocal microscopy immunofluorescence of oocytes
(4% SDS, 100 mM Tris/HCl, pH 7.5, 0.1 M dithiothreitol, total
The immunofluorescence and imaging protocol was described
volume 70 ml), as described (We used the
previously (The primary antibodies
spike-in SILAC technology combined with high accuracy MS
purchased from Santa Cruz Biotechnology (Heidelberg,
) to quantify age-related proteome changes
Germany) were anti-STAG1 (sc-54466), anti-OCT4 (sc-8628),
in mouse oocytes.
anti-ZAR1 (sc-55994) and anti-BUB1 (sc-18286). Appropriate
The spike-in method employs an independently prepared
Alexa Fluor-tagged secondary antibodies (Invitrogen, Life
internal or spike-in standard (in our case the F9 EC cells, heavy)
Technologies, Darmstadt, Germany) were matched to the
against which the test proteome (the oocyte, light) is measured
primaries (Alexa Fluor 488 Donkey Anti-Mouse, A-21202;
after mixing them in a 1:1 ratio. The mixtures were processed
Alexa Fluor 568 Donkey Anti-Goat, A-11057; Alexa Fluor 647
by the FASP–SAX procedure
Donkey Anti-Rabbit, A-31573). The fluorescence signal
The peptide fractions obtained from the FASP–SAX procedure
intensity was quantified using Image J (U.S. National Institutes
were analysed by LC-MS/MS on a LTQ Orbitrap Velos
of Health, Bethesda, MD, USA).
mass spectrometer (Thermo Scientific, Waltham, MA, USA),
Reproduction (2014) 148 55–72
C Schwarzer, M Siatkowski and others
equipped with an Easy nano-LC system and a nano electrospray
Accuracy and reproducibility of our SILAC approach
source (both from Proxeon, Odense, Denmark). Briefly,
We applied two controls to validate our spike-in SILAC
peptides were separated by reversed-phase chromatography
approach. These controls are based on the ‘ratio-of-ratios'
on fused silica capillary chromatography columns (15 cm
method (In the first control, we examined
length, ID 75 mm; New Objectives, Inc., Woburn, MA, USA)
the heavy:light ratio distribution of F9 cells and the heavy:light
that were packed in-house with Reprosil pure C18 material
ratio distributions of F9 cell/oocyte mixtures. The latter were
(3 mm; Dr Maisch, Ammerbuch, Germany). We used gradients
markedly broader than the former (judged by the number of
from 2 to 27% of buffer B (80% acetonitrile and 0.5% acetic
standard deviations, S.D., into which 90% of the values fall; see
acid) for the SAX flow through fractions, for the pH 11 washing
below), confirming the expected larger dissimilarity of the F9/
step and for the first elution at pH 8; from 5 to 35% B for SAX
oocyte mixture over the F9/F9 mixture (
elution steps at pH 6 and 5; and from 8 to 40% B for the SAX
In the second control, we examined the ‘ratio-of-ratios'
elution steps at pH 4 and 3. Each gradient lasted 190 min and
distributions of the samples of pubertal and climacteric oocytes
was followed by a gradient over 10 min to 90% B and further
relative to mature age oocytes and of mature age replicates
elution at 90% B for 5 min before the column was equilibrated
against each other. To this end, we harvested one pool of 1400
again with starting buffer A (0.5% acetic acid). The mass
oocytes from mature mice (collected from 47 mice all on the
spectrometer was operated in data-dependent mode (positive
same day; note that the number of climacteric mice needed for
ion mode, source voltage 2.1 kV), automatically switching
an equivalent experiment would be around six times larger)
between a survey scan (mass range m/zZ350–1650, target
and split it into two mature age replicates. The protein data
valueZ1!106; resolution RZ60 K; lock mass set to back-
associated with the mature age replicates are available from the
ground ion 445.120025) and MS/MS acquisition of the 15 most
PRIDE archive. We provide the interval (number of S.D from the
intense peaks by collision-induced dissociation in the ion trap
zero centre) into which 90% of the ratio values fall as a measure
(isolation width m/zZ2.0; normalized collision energy 35%;
of how broad or narrow the distributions are. These are 9.957
dynamic exclusion enabled with repeat count 1, repeat
S.D. for the pubertal vs mature oocytes, 10.189 S.D. for the
duration 30.0, exclusion list size 500 and exclusion duration
climacteric vs mature oocytes, but 4.286 S.D. for the mature age
set to 90 s; double charge and higher charges were allowed).
replicates (These resultsindicate that diversity between the different age groups is larger
Protein identification and quantification
than within the same age group – a prerequisite to analyse theoocytes of different maternal ages comparatively. Unless
Raw data were processed by MaxQuant Software (v 1.2.2.5,
otherwise stated, the mature age group was used as a reference
Martinsried, Bavaria, Germany) involving the built-in Andro-
to analyse the variation and concordance of protein and
meda search engine. The search was performed against the
transcript levels during maternal ageing.
International Protein Index database (mouse IPI version 3.73;concatenated with reversed sequenceversions of all entries and supplemented with common
Bioinformatics and statistical data analysis
contaminants. Parameters defined for the search were trypsin
The microarray raw data were imported into the R environment
as the digesting enzyme, allowing two missed cleavages; a
(We quantile-normalized the data and
minimum length of six amino acids; carbamidomethylation at
filtered out probe sets of a low signal level using the
cysteine residues as fixed modification, oxidation at methion-
Agi4x44Preprocess package (Bioconductor;
ine and protein N-terminal acetylation as variable modifi-
) with default parameters. Expression values of probe sets
cations. The maximum allowed mass deviation was 20 ppm for
mapping to the same gene were averaged, yielding 22 334
the MS and 0.5 Da for the MS/MS scans.
unique entities. The correlation of mRNA and protein
Protein groups were identified with a false discovery rate set
abundance values was calculated by Kendall rank (non-
to 1% for all peptide and protein identifications separately,
parametric) t correlation coefficients.
when there were at least two matching peptides, at least one of
In the proteome data provided by the MaxQuant Mass
which was unique to the protein group. Primary quantification
Spectrometer Data Analysis Software, proteins are placed in
was performed using the heavy F9 lysate mix as an internal
one group if the identified peptide set of one protein is equal to
standard, and ratios between corresponding heavy and light
or contained in a second protein's peptide set. Then, the
peptide versions were normalized to this mix and expressed as
proteins with the highest peptide count in a group are retained.
H/L (i.e. heavy/light SILAC internal standard/sample). All these
The IPI (updated to release 3.87) identifiers of these proteins
primary protein ratios are the means of at least two (heavy and
were mapped to ENTREZ identifiers with an existing MGI
light) peptide ratios. Direct comparison of oocyte samples was
symbol. We obtained a single ENTREZ identifier for 96.22% of
achieved by a ‘ratio-of-ratios' calculation, which is possible
the protein groups to which we assigned the logarithm (to base 2)
because the internal standard was the same for all samples
of the heavy:light ratio calculated by MaxQuant. Ties that may
occur due to isoforms (splice variants) or close paralogs that are
The MS proteomics data have been deposited into the
not distinguishable based on the peptides identified are
ProteomeXchange Consortium (
handled by downweighting the logarithmic heavy:light ratio,
via the PRIDE partner repository (
defining the base b of the logarithm as bZ2C(TK1)/T, where
with the dataset identifier PXD000512.
T is the number of ties. Thus, proteins with ties have lower
Reproduction (2014) 148 55–72
SILAC proteome analysis of mouse oocyte ageing
influence in subsequent analyses. In the case of proteins
we refer to this algorithm as the ‘elimination
mapping to the same ENTREZ identifier, we average the
algorithm'. Finally, GO term heat maps were generated with a
logarithmic heavy:light ratios. We subjected the proteins
P value cut-off of 0.01, and a gray gradient corresponding to the
identified to further analysis using a ‘ratio-of-ratios' (
P value from light gray (slight overrepresentation, high P value)
) describing the ‘age-transition-based expression
to dark gray (marked overrepresentation, low P value), using
change' from pubertal to mature and from mature to
white as default (no overrepresentation).
climacteric. Specifically, we subtracted the log2 F9:oocyteratio of the mature age group from the log2 F9:oocyte ratioof the pubertal group, and we subtracted the log2 F9:oocyte
ratio of the climacteric group from the log2 F9:oocyte ratioof the mature age group.
Maternal ageing of oocytes features both deteriorationand improvement of functional properties
Correlation analysis of transcriptome and proteome
In order to give our proteomic analysis a phenotypicfoundation, we firstly obtained a functional portrait
We asked whether maternal ageing within the proteome–
of the MII oocytes ovulated by B6C3F1 mice aged
transcriptome intersection causes regulated changes in the
3 weeks (pubertal), 8
proteome of oocytes compared to their transcriptome. We
G1 weeks (mature) and 58G10
considered that the analytical methods of LC-MS/MS and
weeks (climacteric). As expected, the number of oocytes
microarray may have different dynamic ranges; we, therefore,
ovulated decreased (and the frequency of
calculated their correlation using a statistical method that is
hyperhaploid karyotypes increased during maternal
independent of the absolute amount of change. The Kendall
ageing: 7% (1/13), 0% (0/39) and 30% (9/30) respec-
tau (t) coefficient of correlation measures the similarity of the
tively. In spite of these negative changes, the proportion
proteome (X) and transcriptome (Y) protein/gene orderings
of oocytes competent for blastocyst formation increased
when ranked by their respective changes of abundance,
with maternal age, as measured by blastocyst rates at
regardless of any threshold. In other words, it measures the
96 h after ICSI (
similarity of the orderings of the data (X and Y) when ranked
Higher cell numbers accompanied the increase in
by each of the quantities. If the agreement between the two
blastocyst rates. The blastocysts' total cell numbers
rankings is perfect (i.e. the two rankings are the same), the
in the ICSI group increased significantly with maternal
coefficient has a value of 1. If the disagreement between the
age (pubertal: 36.7G13.6 cells, nZ32; mature: 45.8G
two rankings is perfect (i.e. one ranking is the reverse
15.7 cells, nZ33; climacteric 55.8G14.2 cells, nZ16;
of the other), the coefficient has a value of K1. If X and Y
ANOVA, PZ0.0001; multiple comparisons using t-test,
are independent, then the coefficient is zero.
In addition to fertilization (ICSI), developmental
competence was also tested by parthenogenesis (PA) and
GO BP overrepresentation analysis
by SCNT from cumulus cells (B). PA lacks
In order to address the concern of a sampling bias of the
contribution from sperm; SCNT eliminates a possible
proteome with respect to the transcriptome, we sampled 1000
effect of meiotic aneuploidy on development. In both
random sets from the transcriptome, of the same size as the
PA and SCNT, maternal age correlated positively with
proteome, and calculated the GO semantic similarity score
an increase in blastocyst rate at 96 h (Fisher's exact test,
between the proteome and the transcriptome and between the
P!0.01). Cumulus cells from a transgenic donor harbour-
random sets and the transcriptome using the GOSemSim
ing green fluorescent protein (GFP) under the control of
the regulatory element of Pou5f1 i.e. Oct4 (Oct4–GFP)
Considering both proteome age transitions, two gene lists
were used for SCNT, which allows the visualization of
ranked by the expression values were subjected to GO BP
pluripotent gene expression. Higher OCT4–GFP intensity,
overrepresentation analysis. Following
which is a predictor of developmental competence
we applied a cut-off-free protocol to each gene list,
(was observed in derivative cloned
following rank square transformation, treating positive and
morulae obtained from the oocytes of older mothers
negative expression changes in the same way. We then
calculated the Mann–Whitney U test statistics for all gene
Z0.0364; multiple comparisons using t-test,
sets that correspond to GO BP terms, investigating whether
PZ0.011 for climacteric vs pubertal age; .
genes in these gene sets rank higher in the transformed ranking
Although it is well established that deterioration of
than the genes not in the gene set. Then, we drew 1000 random
genome stability (e.g. meiotic aneuploidy) is a hallmark
sets of the same size from all genes in the detected proteome,
of oocyte ageing, our data attest to the presence of other
for each GO BP term. Again using the transformed ranking, we
features that clearly do not deteriorate with maternal
calculated the Mann–Whitney U test statistics for the random
ageing, as revealed by improved blastocyst formation,
sets, thus obtaining P values describing the significance of the
blastocyst cell numbers and Oct4 transgene expression.
GO term enrichment. We also applied an algorithm in
Our aim here is to shed light on how oocytes age in
processing the ontology terms that returns more specific GO
molecular terms. The variable rates and qualities of
BP terms by considering the dependencies between them
blastocyst formation document that the nature of the
Reproduction (2014) 148 55–72
C Schwarzer, M Siatkowski and others
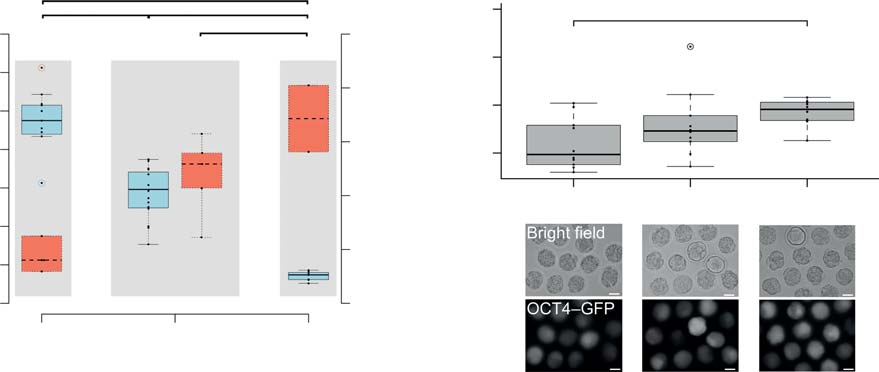
OCT4–GFP intensity
Number of oocytes o
Developmental rates to blastocyst obtained from pubertal, mature age and climacteric oocytes after SCNT, ICSI and PA.
28 (9.2±3.4)†*
63 (19.1±32.3)†*
156 (32.8±31.1)†*
160 (38.6±4.2)†*
132 (44.6±14.8)†*
166 (69.5±7.4)†*
Climacteric (58±10)
126 (48.8±5.4)†*
95 (72.0±17.5)†*
†Fisher's exact test, pairwise comparison related to the one-cell stage and within groups (e.g. SCNT), *P<0.01.
n, number of replicates.
Figure 1 Increased developmental potential of oocytes ovulated from B6C3F1 mice of advanced maternal age. (A) Numbers of oocytes ovulated inassociation with blastocyst rates after ICSI in pubertal, mature and climacteric mice. Significance was tested by ANOVA, pairwise comparison wasmade with a multiple comparison t-test (*P%0.05). (B) Blastocyst rates observed after fertilization (ICSI), parthenogenesis (PA) and cloning (SCNT)given as both total (sum over replicates) and meanGS.D. Significant differences in blastocyst rates between age groups were compared using pairwiseFisher's exact test. (C) Bright field and fluorescent images of morulae after SCNT from OG2F1 cumulus cells (transgenic for Oct4-GFP); size bar,50 mm. Significance was tested by ANOVA (*P%0.05).
starting material – the oocytes – was not constant during
respectively 140 female mice were required in the
maternal ageing, beyond the notorious phenotype of
climacteric group to obtain 700 oocytes. Given the
meiotic aneuploidy. The features of maternal ageing may
scarce amount of material, especially in the climacteric
be rooted in the nucleus or in the cytoplasm of the
group, we adopted a spike-in (SILAC) quantitative
oocyte. While these cellular components are physically
method for proteome analysis, coupled to high-
mixed in MII oocytes (absence of nuclear envelope),
resolution liquid chromatography combined with MS
they can be disentangled in silico by performing GO
(LC-MS/MS). We used F9 cells labelled isotopically with
analysis of gene expression data in the domain ‘cellular
heavy lysine (Lys8) and heavy arginine (Arg10) as
component'. We considered that since proteins relate
material for the spike-in. Oocyte lysates were mixed
more closely to cell phenotypes than mRNAs, protein
1:1 to lysates of F9 cells that had been labelled efficiently
analysis may reveal aspects of the maternal age effect
(97.8%; Thus, the proteins
that could not be grasped so far by transcript studies.
detected in both oocytes and F9 cells are quantifiablerelative to each other in this study. Using a linear ion-traporbitrap hybrid mass spectrometer (LTQ Orbitrap) and
A SILAC screen of the proteome of MII mouse oocytes
the MaxQuant Proteomic Software, we were able to
of different maternal age
identify a total of 3268 different protein groups, of which
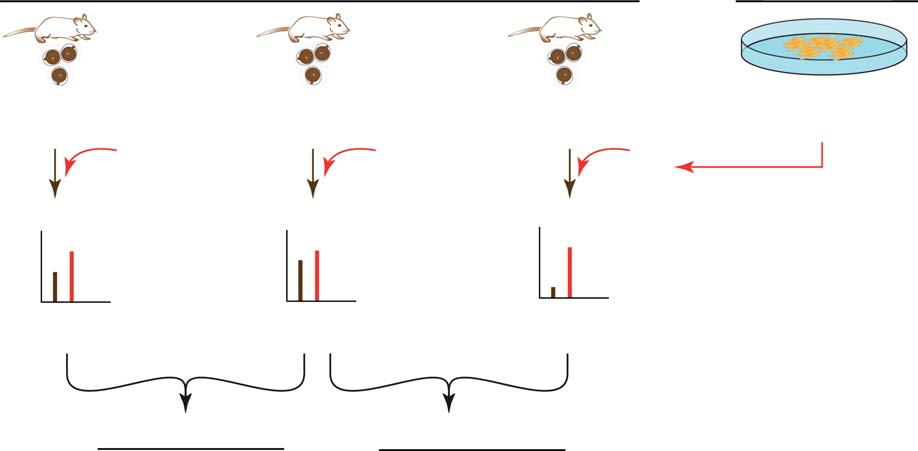
Seven hundred zona-denuded MII oocytes were used for
2654, 2639 and 2617 groups were found in pubertal,
each of the three age groups (3 weeks, pubertal; 8G1
mature and climacteric MII oocytes respectively
weeks, mature age and 58G10 weeks, climacteric; see
(pubertal h mature, 2450 groups and mature h
for a graphical visualization of the SILAC
climacteric, 2451 groups). Here, each protein group
experimental set-up). While 15 and 24 female mice
identified is defined by an F9:oocyte ratio (heavy:light
were sufficient in the pubertal and mature age groups
isotopic ratio). Accuracy and reproducibility of our SILAC
Reproduction (2014) 148 55–72
SILAC proteome analysis of mouse oocyte ageing
approach were validated (see
that may occur because of isoforms/splice variants (see
and ‘Materials and methods' section).
‘Materials and methods' section for details), mapping
The protein groups identified were mapped to ENTREZ
resulted in 2773 gene identities for the pubertal age
identifiers with an existing MGI symbol. Owing to ties
group, 2757 for the mature and 2732 for the climacteric
with Lys8 and Arg10
Non-labelled oocyte samples of different age groups (light)
ratio F9:oocyte (heavy/light)=
quantified protein level
2 F9:oocyte (pubertal)
2 F9:oocyte (mature age)
Log2 F9:oocyte (climacteric)
F9 oocyte (pubertal)
F9 oocyte (climacteric)
Age transition-based
expression change:
F9 oocyte (mature age)
F9 oocyte (mature age)
=> Proteins changing during first transition => Proteins changing during second transition
Simultaneously detected
(8±1 weeks) (58±10 weeks)
in all age groups
No. of transcripts (microarray)
No. of detected protein groups
No. of corresponding
gene identities (proteins)
– without probe on microarray: 140– with corresponding transcripts below detection limit:
No. of gene identities in
No. of gene identities (proteins)
detected in age transitions
(first age transition)
(independent of transcript detection)
(second age transition)
No. of gene identities (proteins) changingmore than four fold in age transitions
(independent of transcript detection)
(first age transition) (second age transition)
(in both age transitions)
Figure 2 Schematic overview of the SILAC workflow and summary. (A) Oocyte lysates were each mixed 1:1 with labelled F9 cell lysate (SILACreference standard, ‘spike-in'). Mixtures were fractionated and analysed by LC-MS/MS. Primary quantification was performed using the heavyF9 cells as SILAC internal standard (*): ratios between corresponding heavy (F9 cells) and light (oocyte) peptide versions were used to determineprotein expression levels (expressed as H/L, heavy/light, i.e. SILAC internal standard/sample). Secondary quantification (age transition-basedexpression change) was performed by dividing the individual H:L peptide ratios of the age groups to be compared (a ‘ratio-of-ratios' calculation).
(B) Summary of the amounts of transcripts/proteins detected, identified and analysed under the specified criteria.
Reproduction (2014) 148 55–72
C Schwarzer, M Siatkowski and others
age group. A summary of all proteins identified and all
and from mature to climacteric (second age transition),
peptides detected with their mass accuracies is provided
using mature as the point of reference. We adopted a
in see section on
fourfold threshold to set significance, because it is in a
given at the end of this article.
large excess of the variation (max. 0.41-fold) shown by
Representative protein spectra are shown in
housekeeping gene products HPRT1, H2AFZ and PPIA
A total of 2324 of these mapped gene
(A) and because twofold
identities were common to the three age groups (B
fluctuations in proteomics can also be seen in technical
and The raw protein data
replicates (Of the 2066 proteins
associated with the three age groups are available at
shared by all age groups and with detected mRNA, 48
the ProteomeXchange Consortium (see link in ‘Materials
and 42 proteins varied greater than or equal to fourfold in
and methods' section).
the first and in the second age transition respectively. The
We performed conventional microarray (Agilent,
mathematical union (g) of these two groups of proteins
Santa Clara, California) analysis of these oocytes in
yields 69 proteins (3% of 2066). In contrast to proteins,
parallel with the proteome analysis. The transcriptomes
the abundance of only one mRNA, namely Cenp-e,
were generated from 20 MII oocytes collected in
changed fourfold during the first age transition and none
biological triplicates for each age group (pubertal,
in the second. CENP-E is a kinesin-like protein
mature and climacteric). A total of 22 334 gene identities
associated with kinetochores. Its mRNA was identified
were common to the transcriptomes of the three age
as age-regulated in mouse oocytes
groups (The raw microarray data associated
Overall, the quantitative change in the oocyte
with this manuscript are available at the Gene Expression
proteome (69/2066 proteins; 3%) exceeded the quan-
Omnibus (see link in ‘Materials and methods' section).
titative change in the protein–matched transcriptome
A 140 of the 2324 gene identities of the proteome had
(1/2066 genes 0.05%) (c2 test, PZ2.46!10K16), and the
no corresponding probe on the microarray (
concordance of the two is essentially nil. The overall lack
of correlation between proteome and transcriptome,
given at the end of this article); cognate mRNA was
calculated on all 2066 genes/proteins, is summarized
below detection level for another 118 gene identities,
by a Kendall t rank correlation coefficient that is not
despite the presence of the probe on the microarray
significantly different from zero: 0.0262 in the first
). Hence, 2066 gene identities
age transition (PZ0.0743) and 0.0053 in the second
were simultaneously detected in the transcriptome and
age transition (PZ0.7184, see also ‘Materials and
proteome in all age groups (transcriptome–proteome
methods' section).
intersection, A summary of thenumbers of proteins detected is provided in
Bioinformatics analysis based on GO terms revealed
The most differently expressed proteins of the ageing
that the two datasets for the proteome and the
oocyte proteome are predominantly localized in thenucleus
transcriptome feature similar BPs. The GO semanticsimilarity (GOSemSim) score is 0.899, which is within
Because quantitative change (greater than or equal to
the interval defined by the lower and upper quartiles
fourfold) in the oocyte transcriptome is not correlated
(0.894 and 0.901 respectively) of the distribution of
with that in the proteome, we decided to consider all
the similarity scores between the random sets and the
proteins detected in subsequent analyses, irrespective of
transcriptome (see ‘Materials and methods' section for
whether they have cognate mRNAs or not. This decision
details on how the GOSemSim was calculated).
resulted in a slight increase in the number of changing
Although the number of proteins identified is smaller
(greater than or equal to fourfold) proteins, from 48 to 55
than the number of transcripts, the proteins we detected
in the first age transition and from 42 to 49 in the second
can thus be considered representative of the complete
age transition. These proteins are listed in along
with the direction (b, increase and a, decrease) ofquantitative change and the GO domain ‘cellularcomponent' with which they are annotated. The two
Quantitative change in the oocyte proteome is not
age transitions (first, puberty to mature age and second,
predictable from cognate mRNAs
mature age to climacterium) will be contrasted with each
The protein analysis conducted so far has been
other in order to identify those protein changes that are
qualitative (presence/absence). In order to appreciate
specific to the old age.
the quantitative impact of maternal ageing on the oocyte
The relationship of the 55 and 49 age-regulated
proteome, we analysed protein abundances, as defined
proteins (to classic features of ageing is
by the ‘ratio-of-ratios' (; see
described hereafter. Roles in spindle assembly, mainten-
and ‘Materials and methods' section). This reflects age-
ance of centrosome integrity and chromosome segre-
transition-based expression changes, which describe the
gation are featured by HAUS7, ACTR1B, BUB1 and TNT.
transition from pubertal to mature (first age transition)
Another classic feature of ageing, namely oxidative
Reproduction (2014) 148 55–72
SILAC proteome analysis of mouse oocyte ageing
First age transition
Second age transition
Figure 3 Expression profiles of selected proteins in B6C3F1 oocytes during maternal ageing. Change in protein abundances of (A) housekeepers,(B) factors related to oxidative stress, (C) culprits of ageing revealed by transcriptome studies, (D) structural maintenance of chromosomes (SMC) andspindle assembly checkpoints (SACs) and (E) maternal-effect factors. Values are expressed as fold-change during first and second age transition.
Reproduction (2014) 148 55–72
C Schwarzer, M Siatkowski and others
Table 1 Proteins undergoing change in expression during the first (puberty to mature age) and second age transition (mature age to climacteric) inexcess of fourfold (in alphabetical order).
First age transition
Second age transition
Cellular component
Cellular component
C, cytoplasmic; N, nuclear; NA, not annotated.
aProteins changing in excess of fourfold in both age transitions.
stress, is represented in SART1 and ERO1LB. These
Other age-regulated proteins have a relationship to
two proteins are very loosely related to oxidative
oocyte quality and include ZAR1 (maternal-effect
stress according to the database of the European
factor), PAPOLA (mRNA maturation) and TCL1B1
Bioinformatics Institute (and
(developmental potential). PAPOLA is the poly(A)
they are not known to play a role in mouse oocytes.
polymerase a, which adds adenosine residues to create
Reproduction (2014) 148 55–72
SILAC proteome analysis of mouse oocyte ageing
the 30-poly(A) tail of mRNAs (). TCL1B1
climacteric oocytes. We have tested and confirmed the
is associated with developmental potential in early
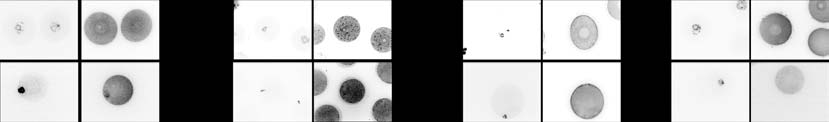
STAG1 profile in situ – directly in MII oocytes of the three
embryos Other proteins that
age groups – using confocal immunofluorescence
changed in abundance include members related to
microscopy and compared the measured intensities.
cytoskeleton assembly/organization (e.g. TMS4BX,
The antibody results matched the LC-MS/MS results
ACTR1B, TUBA3A, MYL1, PTK7, EML4, PLEC, PTK2
(and A0; pubertal vs mature age, P!0.0001;
and JUP) and to protein modification (e.g. CRNKL1,
climacteric vs mature age, P!0.0001; Dunnett's test).
MDN1, MPHOSPH10, DENR and TRIM71).
While the abundance of MAD2L1 and other proteins
Inspection of the GO domain ‘cellular component'
of spindle assembly and chromosome segregation varied
reveals that the proteins in the second age transition that
less than twofold, another component of the spindle
increased with ageing are mainly cytoplasmic, while
checkpoint, BUB1 (), varied
those that decreased are mainly nuclear ; c2 test,
more than sevenfold (As with STAG1, BUB1
PZ0.017). This allocation is not significantly different
abundance was tested by immunofluorescence analysis
from 1:1 in the first age transition (c2 test, PZ0.375).
(B and B0), but it could not be confirmed. This
Among the 49 proteins changing in the second age
discrepancy between SILAC and immunofluorescence
transition, 24 also change in the first transition, while 25
data is discussed below.
changed only in the second age transition. Reflecting
A maternal-effect factor, ZAR1, is among the most
findings already described above, these 25 proteins have
differently expressed proteins of the ageing oocyte
a prevalence of cytoplasmic terms among the increasing
proteome (We also examined the other
proteins and of nuclear terms among the decreasing
maternal-effect factors. Of the 27 maternal-effect
proteins (c2 test, PZ0.054). PAPOLA and ZAR1 are
proteins known (17 were detected in
among the latter ().
oocytes of all three age groups MATER (NLRP5),the maternal antigen that embryos require, is para-digmatic of the maternal-effect factors
Proteins of known candidate genes in the ageing oocyte
and was detected together with the three other members
of the subcortical maternal complex (SCMC), namely
We were surprised in our threshold-based analysis
OOEP (FLOPED), KHDC3 (FILIA) and TLE6 (
(that, except for BUB1, the culprits of oocyte
Abundance of these factors was stable during
ageing were missing, such as BCL2 and BAX (mito-
maternal ageing, as was the abundance of other
chondrial function and apoptosis;
prominent maternal-effect factors, e.g. STELLA (DPPA3)
APACD, SOD1, TXN1 (oxidative stress;
and OCT4, but not ZAR1. ZAR1 abundance declined in
oocytes of climacteric females, as confirmed in situ using
confocal immunofluorescence and comparison of the
NUMA1, SMC1B (spindle assembly and chromosome
measured intensities (and C0; Dunnett's test for
integrity/stability; ,
ZAR1 pubertal vs mature age, PZ0.6108 and climac-
and DMAP1, DNMT1, DNMT3A,
teric vs mature age, PZ0.0043). OCT4 abundance
HDAC1/2 (epigenetic modification; ,
increased slightly in oocytes of climacteric females, but
). Therefore, we searched for them
immunofluorescence analysis revealed a small yet
directly, disregarding the fourfold threshold. APACD,
significant decline in OCT4 levels (Dunnett's test,
BRCA1, DMAP1 and SMC1B were not detected among
P%0.0218; D and D0). These discrepancies
the candidates, while the ones that were detected varied
between SILAC and immunofluorescence data are
less than twofold, except BAX, HDAC1 (and
discussed below.
BUB1 In contrast to these known culprits ofoocyte ageing, three as yet uncharacterized proteins
Molecular signature of oocytes during maternal ageing
loosely related to oxidative stress were found to changein excess of twofold, namely ERO1LB, SART1 and
The most differently expressed proteins of the ageing
glutathione S-transferase omega 1 (GSTO1; B).
oocyte proteome were, only in part, those we had
We continued our analysis focusing on the proteins
expected based on mRNA studies published. In previous
responsible for genome stability (e.g. ploidy) and
transcriptome studies, only a small number of GO BP
embryonic genome activation (e.g. maternal-effect
were affected in old oocytes, based on thresholds that
factors). While SMC1B was not detected in our study,
were sometimes as low as 1.4- to 1.5-fold (
two other members (SMC1A and SMC3) of the structural
maintenance of chromosomes (SMC) complex, which
set out to perform GO analysis to discover the signature
holds the sister chromatids together, were detected in
of the oocyte proteome during maternal ageing.
oocytes of all three age groups, but their abundance
We performed GO overrepresentation analysis using
varied less than twofold D). By contrast, the SMC
ranks instead of age-transition-based expression changes
cofactor STAG1 decreases in excess of twofold in
so that the conclusions of the GO analysis are valid
Reproduction (2014) 148 55–72
C Schwarzer, M Siatkowski and others
Intensity (arbitr
Intensity (arbitr
Intensity (arbitr
Intensity (arbitr
Figure 4 SILAC abundance of selected oocyte proteins validated in situ by immunofluorescence. (A, B, C and D) Representative pictures ofimmunofluorescence staining of STAG1, BUB1, ZAR1 and OCT4 on GV and MII stage oocytes, with DNA counterstaining (YOPRO-1, Life Technologies,Darmstadt, Germany). (A0, B0, C0 and D0) Results of the quantification of the immunofluorescence signal in MII oocytes, performed with Image-J Software.
Significance was tested by ANOVA, pairwise comparison was made with the Dunnett's test, taking the mature age as reference (*P%0.05).
irrespective of any threshold that we may set, such as
fourfold (see ‘Bioinformatics and statistical data analysis'
Although the oocytes ovulated by older mice complied
section for details). We further applied the ‘elimination
with the expected reduction in number and increase of
algorithm' () to account for redundant
meiotic aneuploidy, other commonly expected changes
GO terms. We considered all the proteins of each age
could not be verified. Oocytes of older mice, for
transition (2450 and 2451 respectively), independent of
example, were superior to younger counterparts at
any threshold and of the detection of the protein itself in
accumulating cells during cleavage, forming blastocysts
the other age transition (and
and expressing Oct4–GFP, irrespective of the develop-
see section on given
mental stimulus (ICSI, PA and SCNT). These obser-
at the end of this article). Our overrepresentation
vations confirm and extend our previous study (
analysis in GO BP terms shows that the two age
Thus, while the well-known fact of maternal
transitions feature different BPs (the larger the
age-dependent deterioration of oocyte ploidy was
enrichment, the more the colour shift to dark gray).
confirmed, similar deterioration is not applicable to all
During the first age transition, processes related to
the properties of oocytes, some of which are preserved or
‘regulation of blood pressure', ‘stem cell maintenance'
even improved. The blastocyst phenotypes are evidence
that the nature of the starting material – the oocytes – is
overrepresented. During the second age transition,
not constant during maternal ageing and that there is
however, the analysis shows overrepresentation of BPs
more to oocyte ageing in vivo than the notorious
associated with ‘RNA processing', ‘mRNA splicing, via
increase of aneuploidy. Using the innovative proteomic
spliceosome', ‘positive regulation of nucleocytoplasmic
tool to appreciate this complexity, we are going to
transport' and ‘heart morphogenesis' (), among
discuss that i) proteome analysis of mouse oocytes may
others. In line with the most differently expressed
not be surrogated with transcriptome analysis and ii) the
proteins, these are not the BPs one would expect based
classic features of ageing may not be transposed from
on the GO analyses of transcriptome data, which feature
somatic tissues to oocytes in a one-to-one fashion.
terms related to, for example, oxidative damage and
So far, the molecular bases of oocyte ageing, e.g.
stress response (
meiotic aneuploidy, have been searched for in the
) and inflammation (
mRNA world. Expectations from mRNAs are now
This discrepancy is also discussed below.
confronted with unprecedented information gained
Reproduction (2014) 148 55–72

SILAC proteome analysis of mouse oocyte ageing
Regulation of blood pressure
Stem cell maintenance
Cellular response to light stimulus
Transmembrane receptor protein tyrosine kinase signaling pathway
Peptidyl tyrosine phosphorylation
mRNA splicing, via spliceosome
Positive regulation of nucleocytoplasmic transport
Heart morphogensis
first age transition
second age transition
High P value
Low P value
First age transition: pubertal to mature ageSecond age transition: mature age to climacteric
Figure 5 Gene ontology (GO) overrepresentation analysis reveals proteomic signature of oocyte ageing. Overrepresentation heatmap of GO BPterms of 2450 and 2451 proteins detected in the first and second age transition respectively. For the heatmap, the two protein lists were ranked byexpression values. Rank square transformation was applied for threshold-free and direction of change (up and down)-independent ranking.
Random testing using Mann–Whitney U statistics was applied to obtain P values (cut-off 0.01). Colour code: gray gradient corresponding to theP value, from light gray (light overrepresentation, high P value) to dark gray (marked overrepresentation, low P value), using white as default(no overrepresentation).
from proteomic analysis, providing us with unexpected
proteome correlation also has been described to be poor
results. Our study revealed the highly dynamic character
when analysing stably growing cell lines that are in a
of the oocyte proteome during maternal ageing in vivo,
steady-state and we deem it, therefore, very unlikely that
whereas proteome and transcriptome were qualitatively
our correlation analyses have been hampered by
similar in GO semantic composition. Overall, there was
technical matters (). Further, it is not
a higher prevalence of proteins changing in excess of
surprising that protein fluctuations are inherently larger
fourfold compared to transcripts (69/2066 vs 1/2066)
than mRNA fluctuations.
and minimal concordance between changes of the 2066
observed that distributions of protein copy number per
pairs of protein and transcript values (Kendall t close to
cell have two to three times (in log10 scale) higher
zero). It should be noted that poly(dT)-primers have been
median and higher variation compared with mRNA.
used for the required pre-amplification step when
Whether these larger fluctuations shed light on the
pursuing oocyte microarray analyses. Therefore, only
maternal age effect in oocytes is crucial.
mRNAs with a poly(A) tail that could effectively be
Climacteric oocytes being hard to come by (140 mice
translated at the time of sampling, i.e. the MII stage
needed for 700 oocytes), we relinquished the replicates
oocyte, have been analysed. This may explain why
and the conventional approach of setting twofold
certain proteins have been detected for which no
thresholds combined with P values in order to make
corresponding mRNA could be identified. It is reason-
the call of significance. Instead, we combined a spike-in
able to assume that these RNAs had existed during
method (SILAC internal reference) with a higher
oocyte maturation but lost their poly(A) tail at the time of
threshold of fourfold change. This threshold is higher
ovulation to mark them for degradation. This fact may
than the difference observed by
also have influence on the mRNA expression levels
in their comparison of ageing in different somatic tissues
detected in MII oocytes in general and may partially
and substantially higher (about ten times) than the
explain the missing correlation between transcriptome
variation observed in housekeeping proteins (0.4-fold
and proteome in oocytes. However, the transcriptome to
variation), which are overall stable. Searching the oocyte
Reproduction (2014) 148 55–72
C Schwarzer, M Siatkowski and others
proteome for a group of changes (e.g. GO terms) that
blastocyst stage as the developmental endpoint. This
would stand out from the background, our SILAC study
confirms and extends our previous study (
revealed several proteins that account for the malfunc-
Although we should not aim to explain the
tion of the chromosomal apparatus, but few that reflect
developmental performances on the basis of the
oxidative stress or damage. As this study is the first of
proteome detected (which is still incomplete, despite
its kind, one has to be careful not to over-interpret data.
the best analytical technology used), we note an
Yet, we want to discuss some individual genes that
interesting age-dependent change in the abundance of
specifically caught our attention. Our SILAC results
two factors that may facilitate, or impede, development.
revealed change in the abundance of the SMC cofactor
The abundance of GSTO1 increased in both the first and
STAG1, as well as in that of the SAC protein BUB1 and
second age transition. Although there is no study of
the histone deacetylase protein HDAC1. This latter
GSTO1 in mammalian oocytes, impaired synthesis of
protein modulates the ability of chromosomes to interact
glutathione is indicted as the main cause for compro-
with spindle microtubules and is depleted in climacteric
mised developmental potential of pubertal mouse
oocytes, thereby jeopardizing proper chromosome
oocytes in mice (hence, the increased
segregation (Unlike STAG1 and
abundance of GSTO1 in old oocytes may facilitate
BUB1, our SILAC results reveal no marked change in
development. Abundance of the cell death inducer BAX
the abundance of the core cohesin factors SMC1A
was high in pubertal oocytes and decreased in the
and SMC3. It has been proposed that the efficiency
first age transition. Induction of Bax gene expression
of oxidative phosphorylation in the ageing oocyte is
by in vitro culture conditions correlates with reduced
degraded by free radical attack (
developmental rates of mouse embryos (
however, neither metabolism- nor oxidative stress-
); hence, the higher abundance of BAX in
associated genes featured substantial change of
pubertal oocytes may explain, at least in part, their lower
abundance in our quantitative proteome data. A change
developmental competence compared with mature-age
of proteins very loosely associated with response to
and climacteric oocytes. Certainly, the possibilities are
oxidative stress, e.g. SART1 and ERO1LB, was detected
not exhausted here, and there may be additional factors
in the first and in the second age transition; however,
whose accumulation (developmental agonists) or
a role of these two proteins has not been described in
depletion (developmental antagonists) in oocytes could
oocytes as of yet.
explain the increase in developmental potential
How reliable are fold-differences of protein abun-
observed during maternal ageing.
dance in the SILAC measurement? We found a similar
Although the proportion of blastocyst formation-
abundance trend between our SILAC data and immuno-
competent oocytes increased with maternal age,
fluorescence analysis for STAG1, but not for BUB1. We
aneuploidy would impair subsequent development.
also examined ‘maternal effect factors', i.e. ZAR1 and
Follow-up of the embryos into post-implantation
OCT4 (), and we confirmed ZAR1, but not
development is beyond the scope of this study and
OCT4. On the one hand, while antibodies are limited in
would not add to what is known already
their ability to detect partially degraded proteins due to
The higher blastocyst potential of older oocytes,
the lack of proper 3D structure, proteomics is only
however, does suggest that other features of ageing
dependent on small stretches of peptide sequence and,
oocytes (including non-nuclear processes) may be less
therefore, a degraded protein might still be detected by
affected by maternal ageing than chromosome stability.
LC-MS/MS and MaxQuant. In this respect, while
The majority of proteins that underwent quantitative
proteomics offers more technical accuracy, immuno-
change in the second age transition are classified as
fluorescence offers more biological relevance. On the
cytoplasmic in the GO domain ‘cellular compartment'.
other hand, there are many antibodies that detect distinct
Among the nuclear proteins not directly associated with
protein isoforms (e.g. OCT4A only), whereas LC-MS/MS
SMC, our data revealed a reduced abundance of the
can, in principle, detect all isoforms (contingent on the
poly(A) polymerase a (PAPOLA) and HDAC1, and an
quality of the database). It is important to note that
increased abundance of TCL1B1 in old oocytes. As the
isoforms were not considered during the SILAC analysis
recruitment of many mRNAs for translation depends on
in this study. The prominent maternal factor OCT4,
the length of their poly(A) tail (the
whose A isoform is dispensable for embryo development
reduction in the abundance of PAPOLA may affect the
varied less than twofold during the age
translation efficiency of mRNAs and could further
transitions. Although OCT4 abundance correlates
exacerbate the already poor correlation between tran-
positively with developmental potential, it is probably
scriptome and proteome. We speculate that, at least for
not a precondition, rather it is an effect (
certain cellular functions, oocytes can react to maternal
ageing by operating a shift in how gene transcripts are
On the functional level, we observed an increase in
used, leading to the protein profiles not matching the
developmental rates with ageing in an in vitro culture
mRNA profiles. PAPOLA is also one of the 118 proteins
environment outside of the aged female, with the
with no detected cognate mRNA, together with HDAC1/2
Reproduction (2014) 148 55–72
SILAC proteome analysis of mouse oocyte ageing
(member of the ‘reprogrammome'; ,
different pattern of gene expression changes than ageing
see section on given at the end of
of somatic organs.
this article; suggesting that the
There is one final issue raised by our study. We have
amount of enzyme for these two factors may not be
characterized the maternal age-effect on oocytes, but what
replenished without de novo transcription. We speculate
is the underlying cause? Perhaps, the answer to this
that this might be a fatal hurdle for SCNT
question has to be searched in the somatic niche of the
embryos because de novo transcription of the Papola
ovary. The role of the niche has been investigated in the
gene is dependent on prior nuclear reprogramming,
testis. Spermatogonial stem cells (SSCs) do not age or age
which is known to be inefficient. TCL1B1, which showed
very slowly, as shown by the successful consecutive
an increase in abundance in old oocytes, is a member of
transplantation of SSC populations from the testes of old
the T cell leukaemia/lymphoma 1 (TCL) family and is
mice to the testes of young recipients. These transplanted
specifically expressed in oocytes and stem cells.
SSCs are preserved long past the normal life span of the old
Importantly, Tcl1 is a known downstream target of
donor, for more than 3 years These
Pou5f1 which is one of the four factors
observations suggest that paternal infertility results from
that reprogramme differentiated somatic cells to become
deterioration of the somatic niche, not from deterioration
pluripotent cells TCL1B1
of the germ cells. Ideally, one would like to perform a
is implicated in the developmental potential of mouse
similar investigation also on the ovary, to test whether the
embryos ). The abundance of TCL1B1
quality of oocytes is preserved beyond the life span of the
is increased almost tenfold in old oocytes, suggesting an
female by transplanting her oocytes to a younger niche. In
explanation for their increased blastocyst rate.
principle, this experiment is feasible
When we examined the molecular signature of the
), but very challenging. Until it is performed,
ageing oocyte proteome, we could not confirm the BPs
we think that the available literature already hints to an
indicted by previous transcriptome studies as also being
effect of the niche on oocyte ageing.
altered in the ageing proteome, even though the
showed that adult mice maintained under 40%
proteome is representative of the transcriptome as judged
caloric restriction (CR) did not exhibit maternal age-related
by GO semantic similarity. Transcriptome studies pointed
increases in oocyte aneuploidy, chromosomal misalign-
at genes involved in mitochondrial function, oxidative
ment on the metaphase plate, meiotic spindle abnormal-
damage and stress responses ),
ities or mitochondrial dysfunction, all of which occurred in
genes associated with protein folding/response to
oocytes of age-matched controls that were fed ad libitum.
unfolded proteins, protein metabolism/metabolism, intra-
As the bulk of oocytes have already formed at the time of
cellular transport and cell cycle as well
CR treatment, and as CR suppresses ovulation (
as stress response and response to oxidative damage
we think that the effect of CR is unlikely to
(. Although these three studies also
depend on the oocytes themselves, rather on their niche.
found other families of genes responsive to maternal
Could the above considerations be extrapolated to
ageing, the number of genes was small and their fold-
humans? Caution should always be exercised in extra-
change was low (sometimes as low as 1.4- to 1.5-fold).
polating, due to species-specific differences in life span,
The new technology of SILAC MS can now inform the
oogenesis, duration of the ovarian cycle and onset of
study of oocyte ageing. The most significant terms of BP in
menopause (). The average life
the oocyte proteome include ‘stem cell maintenance' in
expectancy of B6C3F1 females is 128 weeks ().
the first age transition and ‘RNA processing' in the second
The climacteric mice used in our study were aged up to 68
age transition. It is tempting to speculate that these terms
weeks, which would correspond to women 44 years old,
are consistent with a ‘memory' of the oogonium (stem
assuming a linear relationship and a human life expect-
cell)-to-oocyte transition (a recent event in ovaries of
ancy of 84 years for females. Thus, the cells of our study
3-week-old mice) and with a change in the mode of
could have been even older than 68 weeks, which is not a
transcript usage at old age respectively. As the MII oocyte
problem with liver, kidney and brain (
is a transcriptionally silent, terminally differentiated non-
but it is a problem with the ovary as it is depleted of
dividing cell, it cannot use the window of opportunity of
oocytes long before the animal dies. Human oogonia
S-phase to impose new epigenetic marks that alter
divide more times than mouse oogonia until they enter
transcription, such as cycling somatic cells; thus, it
meiosis and these divisions
changes its phenotype through post-transcriptional
may introduce DNA mutations. Perhaps more importantly,
actions. This may partly explain why the GO terms
if we follow up on the effect of CR in mice, then we should
commonly found enriched in ageing somatic tissues (e.g.
consider that the basal metabolic rate per gram of body
inflammation) did not characterize the ageing oocyte
weight is seven times greater in mice than in humans
proteome. This observation is in line with the conclusion
(. Therefore, the human niche may have a
of who compared the ageing ovary
different, i.e. slower rate of ageing than the mouse niche.
and the ageing testis with ageing somatic tissues of mice,
The net outcome of a slower rate of ageing over a longer
finding that ageing of germ cells generally shows a
period of time (decades) is difficult to predict.
Reproduction (2014) 148 55–72
C Schwarzer, M Siatkowski and others
Taken together, our data indicate that ageing of oocytes
Akan P, Alexeyenko A, Costea PI, Hedberg L, Solnestam BW, Lundin S,
on the protein level generally shows a different pattern of
Hallman J, Lundberg E, Uhlen M & Lundeberg J 2012 Comprehensiveanalysis of the genome transcriptome and proteome landscapes of three
gene expression changes than ageing on the mRNA level,
tumor cell lines. Genome Medicine 4 86. ()
and the former does not resemble somatic ageing. The
Alexa A, Rahnenfuhrer J & Lengauer T 2006 Improved scoring of functional
possibilities of modern proteomics are far from exhausted
groups from gene expression data by decorrelating GO graph structure.
(e.g. extended protein coverage, detection of post-
Bioinformatics 22 1600–1607. )
Bouniol-Baly C, Hamraoui L, Guibert J, Beaujean N, Szollosi MS &
translational modifications) and future work will contri-
Debey P 1999 Differential transcriptional activity associated with
bute to a sharper definition of the maternal age effect in
chromatin configuration in fully grown mouse germinal vesicle oocytes.
oocytes, including, but not limited to, the correspondence
Biology of Reproduction 60 580–587. (
between protein immunodetection and LC-MS/MS results.
Cao S, Guo X, Zhou Z & Sha J 2012 Comparative proteomic analysis of
proteins involved in oocyte meiotic maturation in mice. MolecularReproduction and Development 79 413–422. (
Cavaleri FM, Balbach ST, Gentile L, Jauch A, Bohm-Steuer B, Han YM,
Supplementary data
Scholer HR & Boiani M 2008 Subsets of cloned mouse embryos and theirnon-random relationship to development and nuclear reprogramming.
This is linked to the online version of the paper at
Mechanisms of Development 125 153–166.
Chandrakanthan V, Li A, Chami O & O'Neill C 2006 Effects of in vitro
fertilization and embryo culture on TRP53 and Bax expression in B6
Declaration of interest
mouse embryos. Reproductive Biology and Endocrinology 4 61. (
The authors declare that there is no conflict of interest that
Demetrius L 2005 Of mice and men. When it comes to studying ageing and
could be perceived as prejudicing the impartiality of the
the means to slow it down, mice are not just small humans. EMBOReports 6 S39–S44. )
research reported.
Eppig JJ & Wigglesworth K 2000 Development of mouse and rat oocytes in
chimeric reaggregated ovaries after interspecific exchange of somaticand germ cell components. Biology of Reproduction 63 1014–1023.
Esteves TC, Balbach ST, Pfeiffer MJ, Arauzo-Bravo MJ, Klein DC, Sinn M &
This study was supported by the priority programme
Boiani M 2011 Somatic cell nuclear reprogramming of mouse oocytes
(Schwerpunktprogramm) no. 1356 of the Deutsche Forschungs-
endures beyond reproductive decline. Aging Cell 10 80–95. (
gemeinschaft (DFG grants BO2540/3-2 and FUE-583/2-2), by the
Max Planck Society and by the BMBF Verbundprojekt ROSAge,
Geiger T, Wisniewski JR, Cox J, Zanivan S, Kruger M, Ishihama Y & Mann M
2011 Use of stable isotope labeling by amino acids in cell culture as a
FKZ 0315892A.
spike-in standard in quantitative proteomics. Nature Protocols6 147–157. ()
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S,
Author contribution statement
Ellis B, Gautier L, Ge Y, Gentry J et al. 2004 Bioconductor: open softwaredevelopment for computational biology and bioinformatics. Genome
C Schwarzer performed isolation and purification of RNA from
oocytes, immunoconfocal embryo imaging and participated in
Grondahl ML, Yding Andersen C, Bogstad J, Nielsen FC, Meinertz H &
data analysis; B Wang helped with the immunoconfocal embryo
Borup R 2010 Gene expression profiles of single human mature oocytes
imaging; M Siatkowski and G Fuellen performed bioinformatics
in relation to age. Human Reproduction 25 957–968.
data analyses; H C A Drexler designed and performed
Hall VJ, Compton D, Stojkovic P, Nesbitt M, Herbert M, Murdoch A &
proteomics measurements; N Baeumer performed RNA proces-
Stojkovic M 2007 Developmental competence of human in vitro aged
sing and microarray measurements; M J Pfeiffer helped with the
oocytes as host cells for nuclear transfer. Human Reproduction 22 52–62.
mouse work; M Boiani performed oocyte collection for
Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA,
proteomic analysis, karyotype analysis, micromanipulations,
Dudekula DB, VanBuren V & Ko MS 2004 Age-associated alteration of
embryo production and participated in data analysis; and
gene expression patterns in mouse oocytes. Human Molecular Genetics
C Schwarzer, M Siatkowski, M J Pfeiffer, G Fuellen and M Boiani
13 2263–2278. (
wrote the paper. All authors read and approved the paper.
Hodges CA, Revenkova E, Jessberger R, Hassold TJ & Hunt PA 2005
SMC1b-deficient female mice provide evidence that cohesins are amissing link in age-related nondisjunction. Nature Genetics 371351–1355. (
Hu T, Liu S, Breiter DR, Wang F, Tang Y & Sun S 2008 Octamer 4 small
The authors thank Annalen Nolte for help with F9 cell culture
interfering RNA results in cancer stem cell-like cell apoptosis. CancerResearch 68 6533–6540.
and Amy Pavlak for editorial assistance. Excellent animal house
Jiao GZ, Cao XY, Cui W, Lian HY, Miao YL, Wu XF, Han D & Tan JH 2013
support from Ludger Recker and Dr Alexandra Bu¨hler is
Developmental potential of prepubertal mouse oocytes is compromised
due mainly to their impaired synthesis of glutathione. PLoS ONE 8e58018.
Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E & Apweiler R
2004 The International Protein Index: an integrated database for
proteomics experiments. Proteomics 4 1985–1988. (
Ackermann M & Strimmer K 2009 A general modular framework for gene
Kocabas AM, Crosby J, Ross PJ, Otu HH, Beyhan Z, Can H, Tam WL,
set enrichment analysis. BMC Bioinformatics 10 47.
Rosa GJ, Halgren RG, Lim B et al. 2006 The transcriptome of human
oocytes. PNAS 103 14027–14032. (
Reproduction (2014) 148 55–72
SILAC proteome analysis of mouse oocyte ageing
Kosubek A, Klein-Hitpass L, Rademacher K, Horsthemke B & Ryffel GU
Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W
2010 Aging of Xenopus tropicalis eggs leads to deadenylation of a
& Selbach M 2011 Global quantification of mammalian gene expression
specific set of maternal mRNAs and loss of developmental potential.
control. Nature 473 337–342. (
PLoS ONE 5 e13532.
Schwarzer C, Esteves TC, Arauzo-Bravo MJ, Le Gac S, Nordhoff V, Schlatt S
Li L, Zheng P & Dean J 2010 Maternal control of early mouse development.
& Boiani M 2012 ART culture conditions change the probability of
Development 137 859–870. )
mouse embryo gestation through defined cellular and molecular
Lopes FL, Fortier AL, Darricarrere N, Chan D, Arnold DR & Trasler JM
responses. Human Reproduction 27 2627–2640. (
2009 Reproductive and epigenetic outcomes associated with aging
mouse oocytes. Human Molecular Genetics 18 2032–2044.
Selesniemi K, Lee HJ, Muhlhauser A & Tilly JL 2011 Prevention of maternal
aging-associated oocyte aneuploidy and meiotic spindle defects in mice
Lord T & Aitken RJ 2013 Oxidative stress and ageing of the post-ovulatory
by dietary and genetic strategies. PNAS 108 12319–12324. (
oocyte. Reproduction 146 R217–R227. (
Ma P & Schultz RM 2008 Histone deacetylase 1 (HDAC1) regulates histone
Sharov AA, Piao Y, Matoba R, Dudekula DB, Qian Y, VanBuren V, Falco G,
acetylation, development, and gene expression in preimplantation
Martin PR, Stagg CA, Bassey UC et al. 2003 Transcriptome analysis of
mouse embryos. Developmental Biology 319 110–120. (
mouse stem cells and early embryos. PLoS Biology 1 E74. (
Ma M, Guo X, Wang F, Zhao C, Liu Z, Shi Z, Wang Y, Zhang P, Zhang K,
Sharov AA, Falco G, Piao Y, Poosala S, Becker KG, Zonderman AB,
Wang N et al. 2008 Protein expression profile of the mouse metaphase-II
Longo DL, Schlessinger D & Ko M 2008 Effects of aging and calorie
oocyte. Journal of Proteome Research 7 4821–4830.
restriction on the global gene expression profiles of mouse testis and
ovary. BMC Biology 6 24.
Mamo S, Gal AB, Bodo S & Dinnyes A 2007 Quantitative evaluation and
Steuerwald NM, Bermudez MG, Wells D, Munne S & Cohen J 2007
selection of reference genes in mouse oocytes and embryos cultured
Maternal age-related differential global expression profiles observed
in vivo and in vitro. BMC Developmental Biology 7 14.
in human oocytes. Reproductive Biomedicine Online 14 700–708.
McGuinness BE, Anger M, Kouznetsova A, Gil-Bernabe AM, Helmhart W,
Takahashi K & Yamanaka S 2006 Induction of pluripotent stem cells from
Kudo NR, Wuensche A, Taylor S, Hoog C, Novak B et al. 2009
mouse embryonic and adult fibroblast cultures by defined factors. Cell
Regulation of APC/C activity in oocytes by a Bub1-dependent spindle
assembly checkpoint. Current Biology 19 369–380. (
Tarin JJ, Gomez-Piquer V, Pertusa JF, Hermenegildo C & Cano A 2004
Association of female aging with decreased parthenogenetic activation,
Miao YL, Kikuchi K, Sun QY & Schatten H 2009 Oocyte aging: cellular and
raised MPF, and MAPKs activities and reduced levels of glutathione
molecular changes, developmental potential and reversal possibility.
S-transferases activity and thiols in mouse oocytes. Molecular Reproduc-
Human Reproduction Update 15 573–585. (
tion and Development 69 402–410.
Tatone C, Carbone MC, Gallo R, Delle Monache S, Di Cola M, Alesse E &
Monti M, Zanoni M, Calligaro A, Ko MS, Mauri P & Redi CA 2013
Amicarelli F 2006 Age-associated changes in mouse oocytes during
Developmental arrest and mouse antral not-surrounded nucleolus oocytes.
postovulatory in vitro culture: possible role for meiotic kinases and
Biology of Reproduction 88 2. (
survival factor BCL2. Biology of Reproduction 74 395–402. ()
Monetti M, Nagaraj N, Sharma K & Mann M 2011 Large-scale phosphosite
Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J &
quantification in tissues by a spike-in SILAC method. Nature Methods
Nelson LM 2000 Mater, a maternal effect gene required for early embryonic
development in mice. Nature Genetics 26 267–268.
Myers DD 1978 Review of disease patterns and life span in aging mice:
Velde ERT & Pearson PL 2002 The variability of female reproductive ageing.
genetic and environmental interactions. Birth Defects Original Article
Human Reproduction Update 8 141–154.
Series 14 41–53.
Nelson JF, Gosden RG & Felicio LS 1985 Effect of dietary restriction on
Vitale AM, Calvert ME, Mallavarapu M, Yurttas P, Perlin J, Herr J & Coonrod S
estrous cyclicity and follicular reserves in aging C57BL/6J mice. Biology
2007 Proteomic profiling of murine oocyte maturation. Molecular
of Reproduction 32 515–522. (
Reproduction and Development 74 608–616. (
Oliveri RS, Kalisz M, Schjerling CK, Andersen CY, Borup R & Byskov AG
Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J,
2007 Evaluation in mammalian oocytes of gene transcripts linked to
Alpi E, Birim M, Contell J et al. 2013 The PRoteomics IDEntifications
epigenetic reprogramming. Reproduction 134 549–558.
(PRIDE) database and associated tools: status in. Nucleic Acids Research
41 D1063–D1069. ()
Pan H, Ma P, Zhu W & Schultz RM 2008 Age-associated increase in
Vogel C & Marcotte EM 2012 Insights into the regulation of protein
aneuploidy and changes in gene expression in mouse eggs.
abundance from proteomic and transcriptomic analyses. Nature
Developmental Biology 316 397–407.
Reviews. Genetics 13 227–232. (
Pfeiffer MJ, Balbach ST, Esteves TC, Crosetto N & Boiani M 2010 Enhancing
Wallace DC & Chalkia D 2013 Mitochondrial DNA genetics and the
somatic nuclear reprogramming by Oct4 gain-of-function in cloned
heteroplasmy conundrum in evolution and disease. Cold Spring
mouse embryos. International Journal of Developmental Biology
Harbor Perspectives in Biology 5 a021220. (
Pfeiffer MJ, Siatkowski M, Paudel Y, Balbach ST, Baeumer N, Crosetto N,
Walther DM & Mann M 2011 Accurate quantification of more than 4000
Drexler HC, Fuellen G & Boiani M 2011 Proteomic analysis of mouse
mouse tissue proteins reveals minimal proteome changes during aging.
oocytes reveals 28 candidate factors of the "reprogrammome".
Molecular and Cellular Proteomics 10 M110 004523. (
Journal of Proteome Research 10 2140–2153.
Rapti A, Trangas T, Samiotaki M, Ioannidis P, Dimitriadis E, Meristoudis C,
Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I & Gao S 2010
Veletza S & Courtis N 2010 The structure of the 50-untranslated region of
Proteome of mouse oocytes at different developmental stages. PNAS 107
mammalian poly(A) polymerase-a mRNA suggests a mechanism of
translational regulation. Molecular and Cellular Biochemistry 340
Wilding M, Di Matteo L & Dale B 2005 The maternal age effect: a
hypothesis based on oxidative phosphorylation. Zygote 13 317–323.
R Core Team 2012 R: A Language and Environment for Statistical
Computing. Vienna, Austria.
Wisniewski JR, Zougman A & Mann M 2009 Combination of FASP and
Ryu BY, Orwig KE, Oatley JM, Avarbock MR & Brinster RL 2006 Effects of
StageTip-based fractionation allows in-depth analysis of the hippocampal
aging and niche microenvironment on spermatogonial stem cell self-
membrane proteome. Journal of Proteome Research 8 5674–5678.
renewal. Stem Cells 24 1505–1511.
Reproduction (2014) 148 55–72
C Schwarzer, M Siatkowski and others
Wu G, Han D, Gong Y, Sebastiano V, Gentile L, Singhal N, Adachi K,
Zuccotti M, Boiani M, Garagna S & Redi CA 1998 Analysis of aneuploidy rate
Fischedick G, Ortmeier C, Sinn M et al. 2013 Establishment of
in antral and ovulated mouse oocytes during female aging. Molecular
totipotency does not depend on Oct4A. Nature Cell Biology 15
Reproduction and Development 50 305–312. (
Yu G, Li F, Qin Y, Bo X, Wu Y & Wang S 2010 GOSemSim: an R package
for measuring semantic similarity among GO terms and geneproducts. Bioinformatics 26 976–978. (
Received 4 March 2014
First decision 19 March 2014
Zhang P, Ni X, Guo Y, Guo X, Wang Y, Zhou Z, Huo R & Sha J 2009 Proteomic-
based identification of maternal proteins in mature mouse oocytes. BMC
Revised manuscript received 26 March 2014
Genomics 10 348. (
Accepted 31 March 2014
Reproduction (2014) 148 55–72
Outline placeholder
Source: https://ibima.med.uni-rostock.de/fileadmin/Institute/ibima/research_dateien/schwarzer_siatkowski_oocyteAging_Rep_14.pdf
Proceedings of the 27th Annual Meeting of the Brazilian Embryo Technology Society (SBTE), August 29th to September 1st, 2013, Praia do Forte, BA, Brazil. Conference abstracts. Use of bovine sex sorted sperm on timed artificial insemination, in vivo and in vitro embryo production programs P.S. Baruselli1, J.G. Soares1, J.N.S. Sales2, G.A. Crepaldi1, A.H. Souza1, K.A.L. Neves1, C.M. Martins1,
Material Safety Data Sheet 1.Product and company identification a) Product Name: (to indicate the same name or code as shown in label) Lithium Cobalt Oxide b) Recommended use of the chemical and restrictions on use: Suitable for Cathode Material of Li-ion Battery c) Manufacturer/Supplier/Distributor Information: Name, Address, Responsible department Company Name: MTI Corporation Address: 860 South 19th Street, Richmond, CA 94804, USA Tel No. : (510)525-3070








