
Cialis ist bekannt für seine lange Wirkdauer von bis zu 36 Stunden. Dadurch unterscheidet es sich deutlich von Viagra. Viele Schweizer vergleichen daher Preise und schauen nach Angeboten unter dem Begriff cialis generika schweiz, da Generika erschwinglicher sind.
Doi:10.1016/j.cell.2006.04.02
The Expanding Cosmos of Nuclear
Receptor Coactivators
David M. Lonard1 and Bert W. O'Malley1,*1Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030, USA*Contact:
[email protected] DOI 10.1016/j.cell.2006.04.021
About 200 coactivators play a central role in promoting gene expression mediated by nuclear receptors. This diverse group of proteins are key integrators of signals from steroid hormones and have been implicated in cancer and other diseases.
associated proteins such as histone
coactivator's final agenda—that is, to
Nuclear receptors (NRs) comprise a
deacetylases enforce a local chro-
see a particular gene expressed as a
superfamily of conserved transcrip-
matin environment that opposes the
mature functional protein.
tion factors that are activated by their
transcription-promoting activities of
After identification of the first
steroid hormone ligands and play
coactivators (such as histone acetyl-
NR coactivator—the steroid recep-
essential roles in diverse biological
transferase). Through their opposing
tor coactivator SRC-1 (Onate et al.,
processes. For example, the estro-
actions, a balance exists between
1995)—it was predicted that there
gen, progesterone, and androgen
coactivators and corepressors that
might be a small family of coactiva-
receptors are important in reproduc-
defines the magnitude and nature of
tors (perhaps five to ten) that carried
tion; glucocorticoid receptors in glu-
responses to NR ligands.
out the bridging role between tran-
cose metabolism and stress; the thy-
scription factor and transcriptional
roid hormone receptor in oxidative
Coactivators and Transcriptional
machinery. There are now ?200 pub-
metabolism; and PPARs in lipid and
lished NR coactivators that work with
energy metabolism (Mangelsdorf et
Initial experiments in yeast pro-
?48 NRs. Of these, ?50–70 have
al., 1995). Coactivators are molecules
duced a picture of coactivators as
been characterized by more than
recruited by ligand bound activated
"transcriptional adaptors" (Ptashne
one laboratory and have been defini-
NRs (or other DNA binding transcrip-
and Gann, 1990). These adaptors
tively shown to be NR coactivators.
tion factors) that elicit enhanced
were predicted to provide a bridge
Clearly, we are far from identifying
gene expression. In contrast to NRs,
between DNA binding transcription
the totality of authentic NR coactiva-
which are structural y conserved,
factors and the general transcription
tors (http://www.nursa.org) or their
their coactivators are diverse, both
machinery. This simple scenario of
specific functions in the cel .
structural y and in the way they con-
coactivator action turned out to be
Transcription is a highly dynamic
tribute to the transcriptional process,
much more complex.
and orderly process involving many
namely through a diverse array of
Coactivators are predicted to have
subreactions (multiple steps of ini-
enzymatic activities such as acety-
many activities in addition to the
tiation, elongation, splicing, and ter-
lation, methylation, ubiquitination,
initiation of transcription, such as
mination). Given that so many NR
and phosphorylation or as chroma-
mRNA transport from the nucleus,
coactivators have been identified,
tin remodelers. NR coactivators are
mRNA translation, and posttransla-
there is certainly no shortage of them
essential effectors of the biological
tional modifications of the synthe-
to participate in the wide variety of
activities of NRs and their ligands
sized protein. That coactivators pos-
transcription subreactions. But why
(Xu et al., 1999). Although the focus
sess stratified actions in the entire
would a cell possess such a cumber-
of this essay is NR coactivators, con-
process of transcription/translation
some transcriptional apparatus? The
ceptual y they work in a manner simi-
reflects the fact that they do not act
answer may lie in the fact that mam-
lar to general coactivators for other
alone but rather as part of multipro-
mals are substantial y more complex
tein complexes. These multisubunit
than organisms such as yeast, worm,
There is little doubt that the coun-
entities, containing many individual
and the fruit fly, which have far fewer
terparts of coactivators, corepres-
enzymatic activities, represent a
NR coactivators. For instance, only
sors, are equal y important to the
complex machine that is able to con-
a single NR coactivator (Taiman/
cell (Glass and Rosenfeld, 2000).
centrate and link diverse enzymes,
dAIB1) has been identified in fruit
Corepressors interact with NRs
and the processes that they regulate,
flies so far.
that are not bound to ligand and
together in one place. In this way, the
Coactivator activity results in par-
repress transcription. Corepressor-
coactivator complex executes the
ticular physiological consequences.
Cell
125, May 5, 2006 2006 Elsevier Inc. 411
degradation system
PGC-1 coactivator is
plays a positive role
expressed when an
in transcription prior
organism needs to
to a subsequent duty
alter its metabolic pro-
in transcription ter-
gram in response to
mination (Reid et al.,
exercise or cold tem-
2003). This theory
peratures (Lin et al.,
2005). Work in mice
the large number of
lacking the coactiva-
coactivators that are
tors SRC-1 and SRC-
E2 and E3 ligases,
2 reveals their impor-
such as E6-AP, RPF-
tance in carbohydrate
1, UbcH7, and p300.
and lipid metabolism
One can envisage
(Picard et al., 2002).
the recruitment of a
Thus, although at first
procession of coacti-
glance they appear to
vators—for example,
act only in transcrip-
during transcription
tional control, coacti-
initiation, there would
vators are impor-
be SRCs and p68;
tant for modulating
chromatin remodel-
the expression of a
ers such as BRG-1
wide array of physi-
and other ATPase-
ological y important
dependent chroma-
groups of genes.
tin remodelers; and histone
A Cacophony of
(histone acetyltrans-
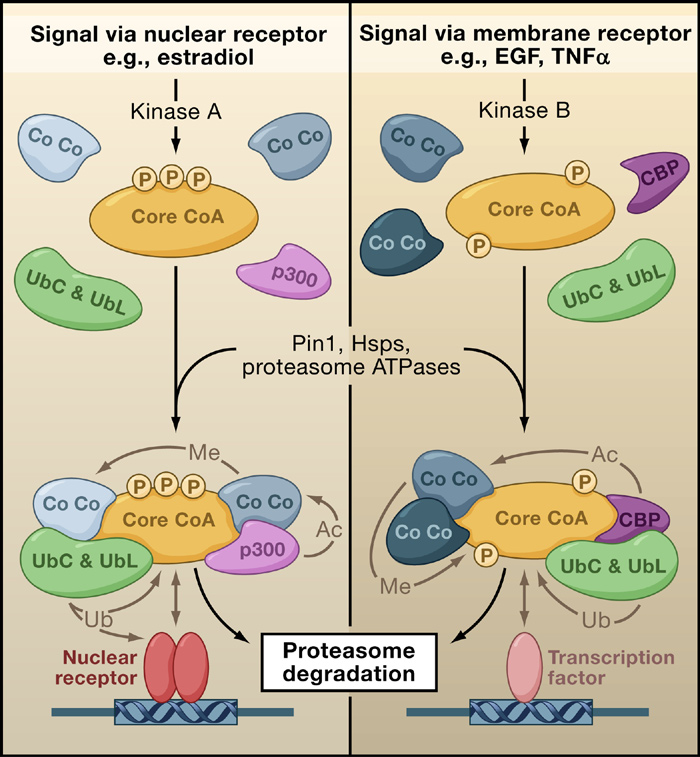
Figure 1. Coactivator-Directed Gene Expression
(Left panel) Activation of a nuclear receptor (NR) by binding of its steroid-hormone
ferases and methyl-
For transcription to
ligand results in activation of kinase A, which induces a distinct phosphorylation
proceed, there need to
pattern (P) in a core coactivator (Core CoA). Protein remodelers (Pin1, Hsps, pro-
as p300/CBP, SRCs,
teasome ATPases) direct the incorporation of a unique set of co-coactivators (Co
be histone modifica-
Co; examples include p300 and the ubiquitin-conjugating ligases UbC and UbL)
and CARM-1. Later
tions (such as acetyla-
into a distinct multiprotein coactivator complex. The composition of this com-
during transcription,
tion and methylation),
plex favors enhancement of NR-mediated transcription of target genes. Various
elongation would be
ATPase-dependent
enzymatic activities assigned to these co-coactivators (acetylation, Ac; methyla-
mediated by P-TEFb
tion, Me) can target protein members of neighboring complexes. This results in
chromatin remodeling,
dissociation of members of these complexes, followed by their destruction by
fol owed by alterna-
initiation of transcrip-
tive splicing of mRNA
tion, elongation, alter-
(Right panel) Signaling through membrane receptors via kinase B induces a phos-
by PGC-1, CAPER,
phorylation pattern distinct from that of Core CoA. This pattern results in the in-
native RNA splicing
tegration of alternative co-coactivators (Co Co), leading to a complex that favors
and CoAA. Final y,
and mRNA process-
transcription mediated by non-NR transcription factors (TF). After their transcrip-
ing, and termination.
tional roles are complete, the complexes are ubiquitinated and degraded by the
plex remodeling or
The focus of coactiva-
the termination of its
tor enzymatic activi-
activities would be
ties in these processes
accomplished by E6-
has centered on the posttranslational
For cessation of transcription, RNA
AP, SSA, and TRIP1 (Metivier et al.,
modification of histones and chroma-
polymerase must dissociate from the
2006). There is no doubt that newly
tin. However, it is becoming clear
gene and reinitiation of transcrip-
identified coactivators will continue
that NRs and their coactivators are
tion must be curtailed. As part of the
to be a prime source for the discov-
also subject to posttranslational
cessation of transcription, coactiva-
ery of new molecular events in tran-
modification. For instance, ligand-
tors and their NRs are modified by
dependent sumoylation of PPARγ
ubiquitination and degraded by the
mediates the repression of inflam-
proteasome. In addition to transcrip-
Coactivators: Integrators of the
matory response genes (Pascual
tional termination, the ubiquitin pro-
et al., 2005). The posttranslational
teasome degradation system is likely
Primary or core coactivators—those
targeting of NRs and their coacti-
to be important in clearing "used"
that interact directly with NRs—exist
vators is important because these
coactivator complexes from the pro-
in steady-state complexes with sec-
modifications influence the expres-
moter, al owing for subsequent steps
ondary or co-coactivator partners
sion of functionally related groups
in sequential transcription to ensue.
(Stal cup et al., 2003) (see Figure
Here, the ubiquitin proteasome
1). The coactivator core complex is
412 Cell 125, May 5, 2006 2006 Elsevier Inc.
composed of a tightly bound invari-
final occupation of binding sites on
Transcriptional Dynamics:
ant group of proteins, whereas the
protein partners is a product of both
Remodeling, Removal, Reinitiation
more loosely bound co-coactivators
their cel ular concentration and their
That many coactivators contribute
associate with the core complex in
affinity for each other. In the case of
to NR-dependent gene expres-
a dynamic, regulated manner. Per-
SRC-3, phosphorylation of a spe-
sion suggests the presence of a
haps a higher-order "complex of
cific combination of residues defines
dynamic force that acts as a pro-
complexes" also forms, enabling
which transcription factors this coact-
pulsion system for these transcrip-
coactivator intercomplex commu-
ivator is able to activate, suggesting
tional machines. In a simple system
nication and efficient integration of
that there may be a "phosphorylation
involving only a few proteins, thermal
signaling pathways such as those
code" (Wu et al., 2005). The selec-
Brownian-driven association and
required for metabolism, growth,
tively phosphorylated coactivator
dissociation would be sufficient to
and inflammation. The fact that
can be conscripted to preferential y
al ow for the necessary proteins to
coactivators belong to distinct com-
implement the expression of genes
associate, perform their enzymatic
plexes may explain how more than
downstream of a particular growth-
roles, and dissociate, al owing other
200 different coactivator proteins
factor signaling cascade. Binding of
proteins to then be recruited to do
individually contribute to cell regula-
coactivators to NRs general y occurs
their jobs. Although Brownian forces
tion in a coherent manner.
through LXXLL interaction motifs in
are likely to play some role, such a
Because coactivators exist as mul-
the coactivator. Many coactivators
simple physical force is unlikely to be
tiprotein complexes, a member of a
possess several different receptor-
sufficient for transcription to ensue.
single coactivator complex can serve
interacting LXXLL motifs, enabling
An orderly procession of coactiva-
as a rate-limiting conduit to control
them to bind to different combina-
tor proteins must associate with the
the actions of the whole complex.
tions of NRs. Complexity is also
promoter for efficient transcription
For example, the phosphorylation
afforded through these LXXLL motifs
(An et al., 2004). Many other pro-
status of SRC-3 defines its associa-
by amino acid residues that flank
teins interact with the promoter in an
tion with other members of the com-
these sequences. In some cases,
orderly sequential fashion (Reid et
plex, such as p300 and CBP histone
these flanking residues are also sub-
al., 2003), such that additional orga-
acetyltransferase or CARM1 meth-
ject to posttranslational modifica-
nizing processes must be involved to
yltransferase (Wu et al., 2005). This
tions, al owing for the dynamic con-
actively disrupt and rearrange these
attendant signaling feature afforded
trol of this NR-interacting motif.
by coactivator complexes suggests
So what are some of the motive
that coactivators may be integrators
Histone Modification: Directed
forces that al ow for orderly remod-
of multiple cell signaling systems.
versus Distributed Regulation
eling capabilities? Protein degrada-
Activation of membrane receptors
Coactivator-mediated histone modi-
tion via the proteasome is one force
and signaling cascades may al ow
fications play an important role in
that makes this procession possible.
the genome to sense the impact of
regulating the transcription of a par-
Because protein degradation medi-
the total environment on the cel .
ticular gene, but the biological impact
ated by the ubiquitin-proteasome
Given that coactivators can organize
is limited usual y to that target gene.
system is a highly regulated and spe-
the expression of "functional groups"
Coactivators, however, can direct
cific process, it is capable of selec-
of genes involved in the execution of
their enzymatic action toward other
tively removing coactivator proteins
a specific regulatory regime (such
coactivator proteins (Xu et al., 2003;
after they have fulfil ed their roles in
as genes involved in metabolism,
Lee et al., 2005). Conceptual y, cross-
transcription, clearing the way for
growth, or cytokine action), they are
posttranslational modification of one
subsequent coactivator associations
prime targets for posttranslational
coactivator by another would al ow the
with the promoter. The ubiquitin-pro-
modification and modulation by
affected coactivator (and the coacti-
teasome system is itself remarkably
kinase cascades (Wu et al., 2005).
vator complex that it resides in) to
complex, as evidenced by the large
Phosphorylation of coactivators by
act in an altered manner on a "global
number of ubiquitin ligases responsi-
kinases, such as IKKα and CDK2,
scale" similar to the far-reaching bio-
ble for the directed targeting of ubiq-
modulates NR-mediated transcrip-
logical effects that kinases exert on
uitin to proteasome substrates, mak-
tion by altering the affinity between
SRC-3 function. Although the histone
ing it the largest class of enzymes
different coactivators and their NRs,
modification code may define the tran-
in mammalian cel s. Another group
influencing which transcription fac-
scriptional state of individual genes,
of proteins that may play essential
tors they are able to coactivate (Wu
coactivator modification codes (acety-
roles in coactivator dynamics are
et al., 2005; Narayanan et al., 2005).
lation, methylation, phosphorylation)
the ATP-driven protein chaperones.
Other modifications, such as methyl-
may define the transcriptional state of
An example of a protein that alters
ation or acetylation, can promote the
broad groups of functional y related
coactivator protein conformation is
dynamic dissociation of coactiva-
genes and may control coactivator
Pin1, a prolyl isomerase that cata-
tor-complex components (Xu et al.,
preferences among NRs and other
lyzes the cis-trans isomerization of
2003; Lee et al., 2005). In the end,
transcription factors (see Figure 1).
proline residues in SRC-3 (see Fig-
Cell 125, May 5, 2006 2006 Elsevier Inc. 413
ure 1). Such a transition in a proline
through transcription of their parent
Lanz, R.B., McKenna, N.J., Onate, S.A., Al-
residue is capable of evoking a large
genes, then those genes also would
brecht, U., Wong, J., Tsai, S.Y., Tsai, M.-J., and O'Malley, B.W. (1999). Cell 97, 17–27.
steric change in the protein and can
require their own set of regulatory
serve as an additional mechanism
coactivators and so on, resulting in
Lee, Y.H., Coonrad, S.A., Kraus, W.L., Jelinek,
to dynamical y alter coactivator pro-
a never-ending circle of regulatory
M.A., and Stallcup, M.R. (2005). Proc. Natl. Acad. Sci. USA 102, 3611–3616.
tein conformation during complex
Coactivators play important roles in
Lin, J., Handschin, C., and Spiegelman, B.M. (2005). Cell Metab. 1, 361–370.
diverse pathological processes, such
RNA Coactivator Complexes
as cancer, inherited genetic diseases,
Mangelsdorf, D.J., Thummel, C., Beato, M.,
SRA is a unique NR coactivator in
metabolic disorders, and inflam-
Herrlich, P., Schutz, G., Umesono, K., Blum-berg, B., Kastner, P., Mark, M., Chambon, P.,
that it is an RNA (Lanz et al., 1999).
mation. The cancer cel , dedicated
and Evans, R.M. (1995). Cell 83, 835–839.
Although a paucity of data exists,
to relentless growth, is certainly a
we believe that there will be other
Matsuura, T., Sutcliffe, J.S., Fang, P., Galjaard,
master at accumulating high levels of
R.J., Yiang, Y.H., Benton, C.S., Rommens,
RNAs that turn out to be NR coacti-
"growth coactivators" such as SRC-
J.M., and Beaudet, A.L. (1997). Nat. Genet.
vators. As for SRA itself, it may play
3/AIB1, thereby assuring a preferen-
15, 74–77.
a structural role as it is subject to
tial rate of expansion (Anzick et al.,
Metivier, R., Reid, G., and Gannon, F. (2006).
a posttranscriptional modification
1997). Germline mutations affecting
EMBO Rep. 7, 161–167.
by pseudouridine synthase (which
E6-AP result in the inherited genetic
converts uracil to pseudouracil) that
Narayanan, R., Adigun, A.A., Edwards, D.P.,
disease Angelman syndrome, (Mat-
and Weigel, N.L. (2005). Mol. Cell. Biol. 25,
alters its conformation (Zhao et al.,
suura et al., 1997). Polymorphisms
2004). RNAs may be an important
in PGC-1 may lead to increased sus-
structural molecule in coactivator
Onate, S.A., Tsai, S.Y., Tsai, M.-J., and
ceptibility to type II diabetes (Lin et
O'Malley, B.W. (1995). Science 270, 1354–
complexes, playing a part similar to
al., 2005). Final y, variations in the
that of structural RNAs in ribosomes.
expression of coactivators among dif-
Pascual, G., Fong, A.L., Ogawa, S., Gamliel,
A number of splicing-related coacti-
ferent individuals may be associated
A., Li, A.C., Perissi, V., Rose, D.W., Wilson,
vators such as p68, p72, CAPER, and
with phenotypic differences among
T.M., Rosenfeld, M.G., and Glass, C.K. (2005).
PGC-1 possess RNA binding motifs;
Nature 437, 759–763.
humans. There is little doubt that we
their uncharacterized RNA binding
have much to learn about the biologi-
Picard, F., Gehin, M., Annicotte, J., Rocchi, S.,
partners may represent other RNA
cal y diverse roles of NR coactivators
Champy, M.F., O'Malley, B.W., Chambon, P.,
coactivators. SRA is also a part-
and Auwerx, J. (2002). Cell 111, 931–941.
and that we have only scratched the
ner in complexes with the SHARP
surface of this expansive coactivator
Ptashne, M., and Gann, A.A. (1990). Nature
and SLIRP corepressor proteins,
suggesting a broader role for RNA
Reid, G., Hubner, M.R., Metivier, R., Brand,
molecules as both coactivators and
H., Denger, S., Manu, D., Beaudouin, J., El-
lenberg, J., and Gannon, F. (2003). Mol. Cell 11, 695–707.
This work was supported by NIH grants to
B.W.O. (HD 08818 and 07857) and NIDDK-
Stallcup, M.R., Kim, J.H., Teyssier, C., Lee,
If coactivators are key regulators of
Y.H., Ma, H., and Chen, D. (2003). J. Steroid
transcription, what regulates their
Biochem. Mol. Biol. 85, 139–145.
expression? Most coactivators appear
Wu, R.C., Smith, C.L., and O'Malley, B.W.
to be regulated either by posttrans-
(2005). Endocr. Rev. 26, 393–399.
An, W., Kim, J., and Roeder, R.G. (2004). Cell
lational mechanisms that determine
Xu, L., Glass, C.K., and Rosenfeld, M.G.
their stability or by control of their
(1999). Curr. Opin. Genet. Dev. 9, 140–147.
activity through phosphorylation,
Anzick, S.L., Kononen, J., Walker, R.L., Azor-sa, D.O., Tanner, M.M., Guan, X.Y., Sauter, G.,
Xu, W., Cho, H., and Evans, R.M. (2003).
although a few, including PGC-1, are
Kallioniemi, O.P., Trent, J.M., and Meltzer, P.S.
Methods Enzymol. 364, 205–223.
regulated through increased tran-
(1997). Science 277, 965–968.
Zhao, X., Patton, J.R., Davis, S.L., Florence,
scription of the parent gene. If regu-
Glass, C.K., and Rosenfeld, M.G. (2000).
B., Ames, S.J., and Spanjaard, R.A. (2004).
lation of coactivators occurred only
Genes Dev. 14, 121–141.
Molecular Cell 15, 549–558.
414 Cell 125, May 5, 2006 2006 Elsevier Inc.
Source: http://retina.umh.es/docencia/confsvivos/articulos/Nuclear%20Receptor%20Coactivators.pdf
HOW TO CREATE A STRUCTURAL SUMMARY Rapid Reference 4.1 Preparing the Transparency Template HOW TO CREATE A STRUCTURAL 1. Photocopy the templates from Appendixes 5a and 5b onto an 8'' x 11'' SUMMARY FOR THE RORSCHACH transparency. Do not enlarge or reduce the size of the form. (It's ofteneasiest to take the book to a full-service photocopy store where you canpurchase a single transparency and have them photocopy it for you.)
Canadian Pari-Mutuel canadienne du pari mutuel Elimination Guidelines © Her Majesty the Queen in Right of Canada, as represented by the Minister of Agriculture and Agri-food Canada, 2016 Catalogue: A22-147/2016 ISBN: 978-0-660-03756-1 CONTENTS INT RODUCT ION………………………………………………….4





