
Cialis ist bekannt für seine lange Wirkdauer von bis zu 36 Stunden. Dadurch unterscheidet es sich deutlich von Viagra. Viele Schweizer vergleichen daher Preise und schauen nach Angeboten unter dem Begriff cialis generika schweiz, da Generika erschwinglicher sind.
Biolchem.huji.ac.il
JOURNAL OF BACTERIOLOGY, Nov. 2005, p. 7518–7525
0021-9193/05/$08.00⫹0 doi:10.1128/JB.187.21.7518–7525.2005Copyright 2005, American Society for Microbiology. All Rights Reserved.
Characterization of Bacterial Drug Antiporters Homologous
to Mammalian Neurotransmitter Transporters
Eyal Vardy, Sonia Steiner-Mordoch, and Shimon Schuldiner*
Department of Biological Chemistry, Alexander Silberman Institute of Life Sciences,
Hebrew University of Jerusalem, Jerusalem, Israel
Received 25 May 2005/Accepted 17 August 2005
Multidrug transporters are ubiquitous proteins, and, based on amino acid sequence similarities, they have
been classified into several families. Here we characterize a cluster of archaeal and bacterial proteins from the
major facilitator superfamily (MFS). One member of this family, the vesicular monoamine transporter
(VMAT) was previously shown to remove both neurotransmitters and toxic compounds from the cytoplasm,
thereby conferring resistance to their effects. A BLAST search of the available microbial genomes against the
VMAT sequence yielded sequences of novel putative multidrug transporters. The new sequences along with
VMAT form a distinct cluster within the dendrogram of the MFS, drug-proton antiporters. A comparison with
other proteins in the family suggests the existence of a potential ion pair in the membrane domain. Three of
these genes, from Mycobacterium smegmatis, Corynebacterium glutamicum, and Halobacterium salinarum, were
cloned and functionally expressed in Escherichia coli. The proteins conferred resistance to fluoroquinolones
and chloramphenicol (at concentrations two to four times greater than that of the control). Measurement of
antibiotic accumulation in cells revealed proton motive force-dependent transport of those compounds.
Multidrug resistance is an increasing problem in antimicro-
stem mainly from difficulties in expressing, purifying, and crys-
bial therapy as well as in treatment of tumors. One of the most
tallizing membrane proteins. One of the approaches suggested
common mechanisms of resistance is removal of toxic com-
for structural studies of low-expression mammalian membrane
pounds from the cell, by drug and multidrug transporters
proteins is to study their bacterial and archaeal homologues.
(8, 26). Multidrug transporters are ubiquitous proteins, and,
Successful examples of this approach can be found in studies of
based on amino acid sequence similarities, they have been
potassium and chloride channels from different sources (4, 5,
classified into several families. Some of these proteins utilize
13, 14). The high-resolution structures of the bacterial homo-
primary energy source and their activity depends on ATP
logues provided an invaluable insight on many aspects of sub-
(ABC transporters) while others utilize secondary energy
strate recognition and transport through biological channels.
sources by coupling their activity to the movement of protons
The high-resolution structures of two bacterial MFS proteins
down a concentration gradient (19, 27).
were solved: LacY, the lactose permease (1), and GlpT, the
The vesicular monoamine transporter (VMAT) catalyzes
phosphate, glycerol 3-phosphate antiporter (10). Although the
the accumulation of neurotransmitters in organelles in ex-
sequence similarity of these proteins is low (⬃20% identity),
change for two protons (34). Besides its known function,
their folds are highly similar. Based on those structures, a
VMAT was shown to protect the cell from the deleterious
model for VMAT was constructed that is in agreement with
effect of toxic compounds by lowering their concentrations in
experimental data (37).
the cytoplasm (18). In addition, the range of substrates recog-
The fold conservation in the MFS proteins and the intrigu-
nized by VMAT is very wide and led to the suggestion that it
ing connection between multidrug transporters of the MFS
behaves as a multidrug transporter (41). Phylogenetic analysis
and the vesicular neurotransmitter transporters led us to iden-
showed that VMAT proteins are evolutionary related to drug
tify and characterize multidrug transporters that are related to
transporters and multidrug transporters of the Major facilita-
VMAT. A BLAST search of the available microbial genomes
tor superfamily (MFS) (32, 34). Like most MFS transporters,
against the VMAT sequence identified several putative pro-
VMAT consists of 12 putative transmembrane segments or-
teins related to VMAT with low but significant similarity
dered in two lobes of six-helix bundles. A feature that distin-
(⬍25% identity). We cloned three of the closest homologues
guishes VMAT proteins from other drug transporters and
and expressed them in
Escherichia coli cells. We report here
multidrug transporters of the MFS is a long loop between
the functional expression and characterization of three new
transmembrane segments TM1 and TM2 that contains glyco-
multidrug transporters from three microorganisms:
Mycobac-
sylation sites (34).
terium smegmatis,
Corynebacterium glutamicum, and
Halobac-
Mechanistic understanding of membrane proteins is limited
by the difficulties in obtaining structural data. These problems
MATERIALS AND METHODS
Sequence analysis. BLAST search using the rVMAT2 sequence as query
* Corresponding author. Mailing address: Department of Biological
against the database of the available microbial genomes was done using the
Chemistry, Alexander Silberman Institute of Life Sciences, Hebrew
NCBI server (http://www.ncbi.nlm.nih.gov/). In order to find more family mem-
University of Jerusalem, Jerusalem, Israel. Phone: 972-2-6585992.
bers the closest sequences derived from the initial BLAST were used for an
Fax: 972-2-5634625. E-mail:
[email protected].
additional BLAST search in the available databases (2). The derived sequences
CLONING OF NOVEL DRUG ANTIPORTERS
TABLE 1. Primers used for cloning the homologues
were compared using ClustalW (36) and a representative dendrogram was drawn
in that ratio in bacteria expressing the putative multidrug resistance (MDR)
using the Njplot software (29). Consensus sequences were detected and pre-
proteins suggests the activity of a proton motive force-dependent efflux system.
sented using the GeneDoc software (25) Hydropathic analysis of the sequences
The ofloxacin accumulation test was carried with 200 l of the resuspended
was done with TMHMM program (38).
culture and was done essentially as described (22) with modifications. After rapid
Bacterial strains and plasmids. E. coli TA15 (7), JM109 (40), C41 (21), and
filtration the filters were incubated for 18 h in 1 ml glycine-HCl buffer (100 mM,
BL21 and HMS174 (Stratagene, La Jolla, Calif.) were used throughout this work.
pH 3), and ofloxacin levels in the buffer were assessed by fluorescence measure-
The pT7-7-Myc-His vector was obtained by removing the
emrE gene from vector
ment using a PerkinElmer fluorimeter (Luminescence Spectrometer LS-50) with
pT7-7-EmrE-Myc-His (23) with restriction enzymes NdeI and EcoRI (New
exciting light at 295 nm and emission at 495 nm. The maximum fluorescence
England Biolabs, Beverly, MA). Homologues of interest were cloned by PCR
signal after addition of CCCP was ⬃110 and it was fitted to accumulation of
using genomic DNA from
H. salinarum,
M. smegmatis, and
C. glutamicum as
36 ng ofloxacin. The chloramphenicol accumulation test was carried with 50 l
templates (provided by M. Mevarech, Department of Microbiology, Tel Aviv
of the resuspended culture and was done by measuring the accumulated
University, H. Bercovier, Hadasah Medical School, Hebrew University of Jerusa-
[3H]chloramphenicol (American Radiolabeled Chemicals, St. Louis, MO) at a
lem, and R. Kraemer, Institute of Biochemistry, University of Cologne, Cologne,
final concentration of 0.5 M chloramphenicol and specific activity of 0.5 Ci/
mmol (200,000 dpm total in the reaction). The maximum signal obtained after
Primers (Table 1) were designed to overlap the ends of the genes and included
addition of CCCP was 35,000 dpm.
sites for restriction enzymes NdeI and EcoRI. The genomes
of H. salinarum and
In both cases, the level of free antibiotics bound to the filter was measured and
C. glutamicum are of high GC content and a successful PCR could only be
subtracted from the antibiotics accumulated in the bacteria. The experiments
achieved using a GC-rich PCR kit (Roche Diagnostics, GmbH, Mannheim,
were carried out in duplicates and repeated at least twice.
Germany) with an annealing temperature of 58°C. Each homologue was cloned
Protein expression. The expression of the three cloned homologues was ex-
into the pT7-7-Myc-His vector. The plasmids obtained were named pT7-7
amined in a strain designed for protein expression:
E. coli HMS174 cells were
HSmdr for the
H. salinarum homologue, pT7-7 MSmdr for the
M. smegmatis
transformed with the three plasmids and expression of the proteins was exam-
homologue, pT7-7 CGmdr for the
C. glutamicum homologue
.
ined. A late-stationar-phase culture was used to inoculate 2XYT medium (33)
Resistance to toxic compounds. Preliminary screens for resistance were done
supplemented with 100 g/ml ampicillin to yield an OD600 of 0.1. The culture
by disk diffusion susceptibility test on inoculated soft agar plates. A 100-l
was incubated at 37°C under aerobic conditions until an OD600 of 0.9, at which
sample of late-stationary-phase cultures
of E. coli JM109 transformed with pT7-
time isopropyl--D-thiogalactopyranoside (IPTG) was added to a final concen-
7(⫺), pT7-7 HSmdr, pT7-7 MSmdr, and pT7-7 CGmdr were used to inoculate
tration of 1 mM. Two hours later, the cells were harvested by centrifugation and
10 ml of warm soft LB-agar (0.7% Agar) (33) that was then poured into plates.
washed once with lysis buffer (150 mM NaCl, 15 mM Tris, pH 7.5, 250 mM
Antibiotic disks (Mast Diagnostics GmbH. Reinfeld, Germany) were placed on
sucrose) before further handling or storage at ⫺70°C.
the soft agar layer. After 18 h, growth inhibition zones, created by the different
Membranes were prepared using lysozyme and hypo-osmolarity for disruption
antibiotics, were compared between the control [pT7-7(⫺)] and the cells express-
of the cells as described (15) except that the volumes used were modified.
ing the homologues. The following antibiotics were examined: amikacin, cefta-
Detection of the protein in the crude membranes was done by Western
zidime, gentamicin, imipenem, meropenem, ofloxacin, tazocin, timentin,
blotting with mouse anti-Myc as a primary antibody and mouse anti-rabbit
ampicillin, cephalothin, colistin sulfate, streptomycin, sulfatriad, tetracycline,
immunoglobulin-horseradish peroxidase as a secondary antibody (Invitrogen
cotrimoxazole, amoxicillin-clavulanic acid, oxacillin, erythromycin, vancomycin,
Carlsbad, Calif.). Detection of the secondary antibody was done with chemilu-
fusidic acid, cefuroxime, nitrofurantoin, ciprofloxacin, and amoxicillin.
minescence Super-signal kit (Pierce, Rockford, IL).
Resistance to chloramphenicol and ofloxacin was studied in more detail in
Protein purification for SDS-PAGE. Membranes were thawed and solubilized
liquid medium:
E. coli JM109 expressing the homologues was grown at 37°C in
in a denaturing buffer (15 mM Tris-Cl pH 7.5, 150 mM NaCl, 2% sodium dodecyl
LB containing ampicillin (100 g/ml) to mid-logarithmic phase to an approxi-
sulfate [SDS], and 6 M urea) at room temperature for 30 min. The solubilized
mate optical density at 600 nm (OD600) of 0.8. The logarithmic cultures were
membranes were then centrifuged (244,000 ⫻ g, 20 min) to remove the non-
diluted to give OD600 of 0.05 and grown in the presence of chloramphenicol or
soluble fraction. The supernatant was incubated with Ni2⫹-nitrilotriacetic acid-
ofloxacin at different concentrations. Growth was assessed by OD600 measure-
agarose beads (QIAGEN, GmbH, Hilden, Germany) for 1 h, with 10 mM
ments after 8 h. Chloramphenicol was dissolved in 100% ethanol to 25 mg/ml.
imidazole. The beads were then washed with denaturing buffer containing imi-
Ofloxacin (LKT Laboratories, St. Paul, MN) was dissolved in sodium acetate
dazole (30 mM). Elution of the protein from the beads was done with sample
buffer (20 mM, pH 4) at a concentration of 4 mg/ml. Both antibiotics were
buffer containing 300 mM imidazole.
diluted in growth medium before adding to the bacterial culture.
Transport of chloramphenicol and ofloxacin. Transport of antibiotics in whole
cells was assessed by measuring their accumulation, essentially as described
before (6). Late-stationary-phase cultures were used to inoculate ampicillin-supplemented LB to an OD
Sequence homology and analysis. The vesicular mono-
600 of 0.02. Bacteria were grown to logarithmic phase,
harvested, and washed once with 50 mM potassium phosphate buffer at pH 7.1.
amine transporter facilitates the accumulation of different
The pellet was resuspended with the same buffer to an OD420 of 20 and kept on
compounds into vesicles using proton motive force. This
ice until assayed: The assay started with 5 min incubation of the cells with 10 mM
protein was shown to belong to the drug/H⫹ antiporters
glucose at 30°C followed by addition of the antibiotics and incubation for
(DHA12) of the major facilitator superfamily. This family is
30 seconds to 10 min. The reaction was stopped by addition of 2 ml of ice-coldbuffer (50 mM potassium phosphate buffer) and rapid filtration through GF/C
divided into five clusters and VMAT is part of a separate
glass microfiber filters (Whatman, Maidstone, England), and after filtration the
branch in one of them (27). A BLAST search of rVMAT2
filters were washed with ice cold buffer.
against the available bacterial genomes (May 2005) revealed
To obtain the equilibration value for all strains, accumulation of ofloxacin and
relatives with low but significant identity to VMAT; the
chloramphenicol was also measured in the presence of 0.5 mM of the protonuncoupler carbonyl cyanide
m-chlorophenylhydrazone (CCCP) for 10 min. The
closest of them had up to 24% identity and
e values up to
ratio between accumulation with and without CCCP was calculated. A decrease
⬃10⫺12. In this BLAST search more than 15 uncharacter-
VARDY ET AL.
J. BACTERIOL.
branches are clearly observed: VMATs, archaeal proteins andproteins from
Corynbacterineae. The
Corynbacterineae branchis divided into two distinct subgroups one from
Corynebacte-rium and one from
Mycobacterium.
Hydropathic analysis of the new sequences revealed, as
usual for MFS proteins, 12 putative transmembrane domainsdivided into two halves by a long cytoplasmic loop betweenTM6 and TM7. In Fig. 2A, a predicted two-dimensional modelof MSmdr is shown.
The multiple sequence alignment of cluster C revealed sev-
eral conserved regions (Fig. 2B) mainly in the N-terminal lobe(TM1, TM2, L2–3, L3–4, TM4, L4–5, and TM5) and to a lesserextent in the C-terminal lobe (L8–9 and TM11). Motifs definedby Paulsen et al. (27) for the entire DHA12 family were com-pared with motifs detected in this cluster. As expected, themotifs are similar between the DHA12 and members of itssubfamily in cluster C but there are some characteristics thatare specific for cluster C: motif D2 is located in TM1 (Fig. 2B).
The most distinct feature of this motif is the two adjacentprolines separated by two residues. In the models created fortwo transporters of this family (37) the conserved prolines arelocated close to the distortion of TM1 suggesting a structuralrole for this motif. Generally, motif D2 is conserved in clusterC as it is in the entire family but the archaeal branch misses thefirst P of the motif (Fig. 2).
Motif A is located in a conserved loop between TM2 and 3
(Fig. 2B) and has been attributed a structural role (reviewed in(27) and (30). In cluster C the motif is hardly changed and itbegins with a highly conserved proline three residues prior tothe conserved glycine. The conserved aspartate downstreamfrom the glycine may be changed in cluster C to glutamate orpolar residues (glutamine and asparagine) (Fig. 2).
FIG. 1. Dendrogram of the main clusters of the DHA12 family.
Motif B is located in TM4 (Fig. 2) and was suggested to be
The tree consists of representatives from each cluster in the DAH12
involved in proton transfer or recognition (28). This RXXXG
family. Cluster A consists of drug and multidrug transporters from
motif is conserved in the whole family and it was shown that a
yeast and fungi. The rest of the clusters consist of sequences fromdifferent bacteria and archaea. The novel multidrug transporters (CG-
mutation in the arginine of this motif resulted in an inactive
mdr, MSmdr, and HSmdr) form differentiated branches in the same
form of the tetracycline transporter, TetAB (11). In cluster C,
cluster with VMAT (cluster C). The sources of the sequences in the
this motif is conserved and expanded at the N terminus with a
dendrogram are as follows. Cluster A: UM05248—
Ustilago maydis,
conserved region that is unique to this cluster (Fig. 2).
Motif C is located in TM5 (Fig. 2) and its main feature is a
maydis, CyhR—
Candida maltosa, and CaMDR1—
Candida albicans.
sequence of three glycines separated from each other by three
Cluster B: Bcr—
Burkholderia mallei, LP_Flo—
Legionella pneumo-
residues (GxxxGxxxG). The spacing between the glycines may
reflect one helix turn and that may point towards a structur-
aeruginosa, CflA—
Coxiella burnetii, EmrD—
Escherichia coli, MdfA—
al role. This motif is conserved in cluster C, and in addition,
Escherichia coli, MDT—
Yersinia pestis biovar Medievalis, and YjiO—
Escherichia coli. Cluster C: rVMAT2—
Rattus norvegicus, hVMAT2—
15 residues upstream from it, there is an additional motif
(C-15) completely conserved in this cluster and absent from
marismortui, HSmdr (YfmO2)—
Halobacterium sp. NRC-1, CEmdr—
other clusters (RgrXXgX) (Fig. 2).
Motif G is located in TM11 (Fig. 2B) and it is a variation of
MSmdr—
Mycobacterium smegmatis, and MAmdr—
Mycobacteriumavium. Cluster D: EF_MEP—
Enterococcus faecium, Bmr—
Bacillus
motif C in TM5, its symmetric counterpart in the N-terminal
subtilis, NorA—
Bacillus cereus, TetA—
Escherichia coli, TetG—
Pasteu-
lobe. In cluster C some additional conserved residues are lo-
rella multocida. Cluster E: CmlA—
Pseudomonas aeruginosa, OpdE—
cated downstream from this motif.
Pseudomonas aeruginosa, LmrP—
Lactococcus lactis.
In the earlier analyses no other variations of motifs were
found in corresponding regions of the DHA12 family (27), butwhen examining the existence of such variants in other TMs
ized, putative proteins with an
e value better than 5 ⫻ 10⫺8
in cluster C, a variation of motif A (TM2) is found in TM8
were detected.
(Fig. 2), and is referred to as motif A2 (Fig. 2B).
Phylogenetic analysis of the putative transporters revealed
Other structural information can be hinted from the sugges-
that they are clustered together with VMAT proteins within
tion of a conserved fold in the MFS proteins. The structural
the DHA12 family (Fig. 1, cluster C). In the figure only some
model of VMAT described an ion pair between the conserved
of the homologues are shown for the sake of clarity but three
aspartate in TM11 and the conserved lysine in TM2 (37). This
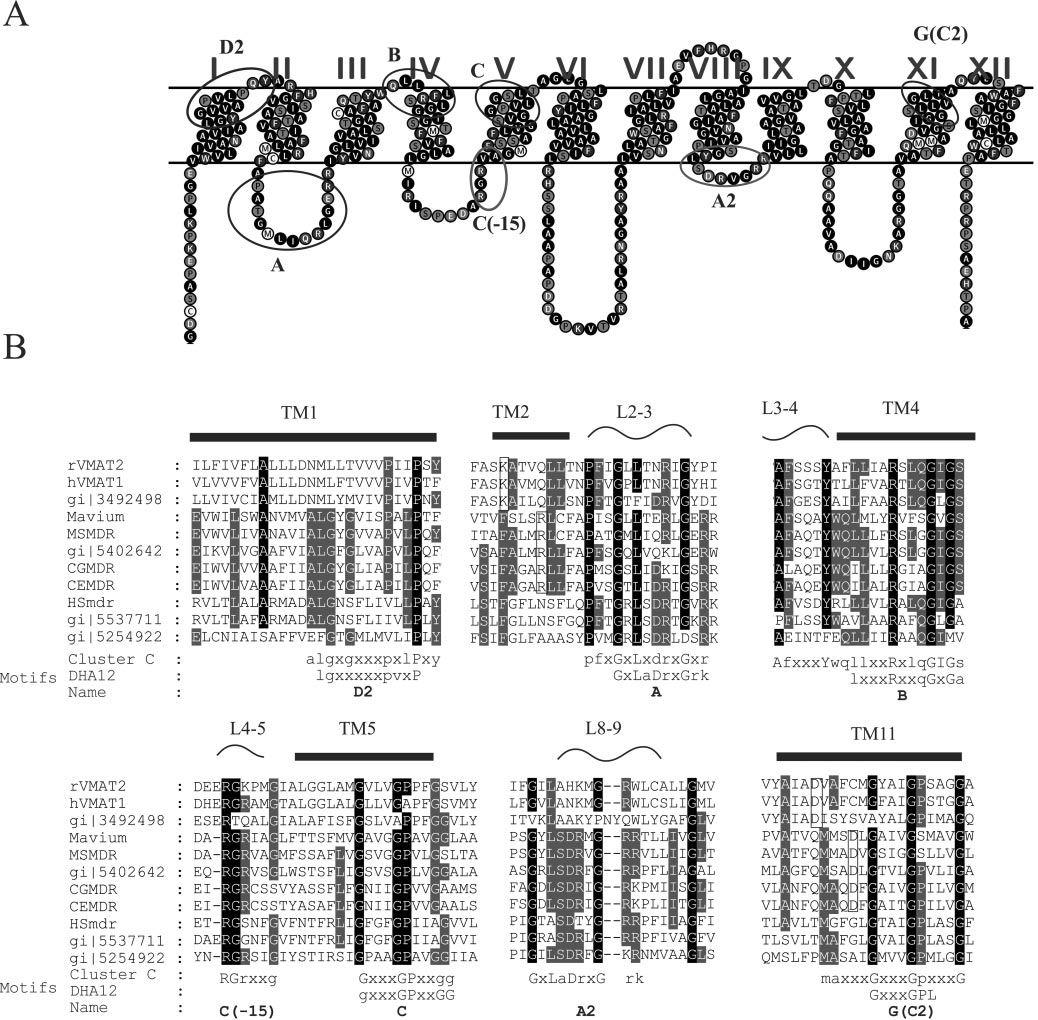
CLONING OF NOVEL DRUG ANTIPORTERS
FIG. 2. Sequence analysis of cluster C. A. A representative topology suggested for DHA12 proteins contains 12 transmembrane domains
divided into two halves by a long cytoplasmic loop between TM6 and TM7. The topology shown is that of MSmdr. Motifs from the DHA12 familyare marked with black circles and motifs specific for cluster C are marked with gray circles. B. Conserved regions in the sequence alignment ofcluster C. At the bottom of the alignment are consensus sequences in which capital letters represent frequency occurrence greater than 90% (darkshading) and small capitals represent frequency occurrence greater than 50% (light gray shading). The consensuses are compared with the DHA12motifs defined by Paulsen et al. (27). The arrows above the sequences point to residues involved in putative ion pairs (open rectangles).
ion pair is supported by experimental data (20). In the Coryn-
charged residues in TM2 and TM11 cannot be buried in the
bacterineae branch this aspartate is shifted one helix turn fur-
membrane without their opposite charge, and as a further
ther towards the C terminus of the TM than in VMAT
support for this notion in the archaeal branch, both charged
(Fig. 2B). Correspondingly, in TM2 an arginine of the Coryne-
residues are absent (Fig. 2B).
bacterineae branch is located exactly one helix turn from the
Representatives from each of the three branches of cluster C
conserved lysine (Fig. 2B). This finding strongly supports the
were cloned and expressed in E. coli: from an unfinished frag-
above proposal for an interaction between the charged resi-
ment of the genome of Mycobacterium smegmatis, MSmdr
dues in TM2 and TM11. This suggestion implies that the
(starting base 2211693); from Corynebacterium glutamicum,
VARDY ET AL.
J. BACTERIOL.
CGmdr (accession number NP_600365); and from the ar-
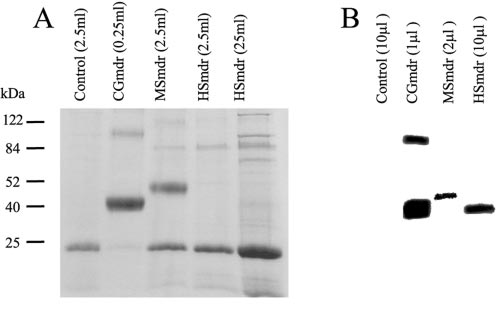
expression level of CGmdr was 20 times higher than that of
chaeon Halobacterium salinarum, HSmdr (also called YfmO2,
MSmdr and ⬃500 times higher than that of HSmdr (Fig. 5A).
accession number NP_279495).
While hardly detectable by Coomassie staining HSmdr from
Phenotype of the novel homologues. The novel DHA12 pro-
the crude membrane fraction was detected by Western blotting
teins were tested for drug resistance. The phenotypic charac-
with an anti-Myc antibody. CGmdr and MSmdr (from 10- and
terization of drug resistance in strains expressing the proteins
5-fold less membrane protein than HSmdr, respectively) were
was done using disk diffusion susceptibility tests with 25 drugs
also detected by Western blot and displayed similar apparent
as described in Materials and Methods. E. coli cells harboring
molecular weights (Fig. 5B). The calculated sizes of the three
plasmids with the homologues exhibited a limited spectrum of
proteins are 42.3, 47.7, and 42.7 kDa for CGmdr, MSmdr, and
resistance (data not shown). Among the compounds tested, the
HSmdr, respectively. As commonly seen for many membrane
most distinct resistance was observed for ciprofloxacin and
proteins, the apparent sizes detected by SDS-PAGE are lower
ofloxacin. The two homologues from the Corynebacterineae
than expected but are consistent with the fact that MSmdr is
(MSmdr and CGmdr) also conferred resistance to chloram-
the largest of the three and the others have very similar mo-
lecular masses. For CGmdr, which is expressed to the highest
To further characterize the phenotype, growth of E. coli
levels, a higher-molecular-weight form that corresponds to a
JM109 cells carrying plasmids with or without the homologues
dimer is apparent in both SDS-PAGE and Western blotting.
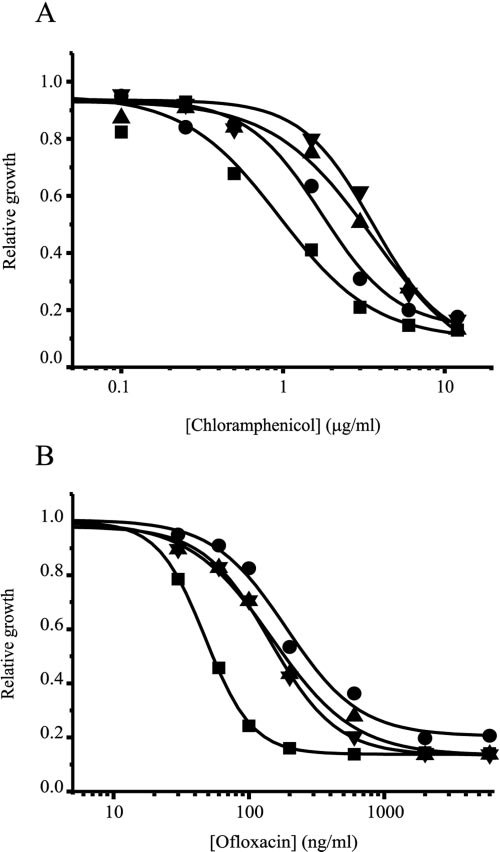
in LB medium containing ofloxacin or chloramphenicol at dif-ferent concentration was examined (Fig. 3A and B). CGmdr-
and MSmdr-expressing strains were equally resistant to bothchloramphenicol and ofloxacin. HSmdr conferred less resis-
In this study, we describe a basic characterization of three
tance to chloramphenicol and slightly more resistance to
proteins encoded by genes belonging to a cluster of the
ofloxacin than the other homologues. In this assay, the approx-
DHA12 family of MFS transporters. A BLAST search of
imate 50% inhibitory concentration (IC ) values for chloram-
the available microbial genomes with VMAT as an input
phenicol are 4 g/ml for CGmdr and MSmdr, 2 g/ml for
yielded several novel, uncharacterized sequences that are
HSmdr, and 1 g/ml for the control (Fig. 3A). The IC
closer to VMAT than any other reported proteins from bac-
for the three homologues is ⬃200 ng/ml, while for the control
teria and archaea. Although the similarity of the new se-
strain it is ⬃50 ng/ml (Fig. 3B).
quences to VMAT is not very high (⬃20%), there are several
Transport of chloramphenicol and ofloxacin by the bacterial
conserved motifs that characterize this cluster. In the DHA12
homologues. Bacteria expressing H⫹/drug antiporters are ex-
family five major clusters have been previously identified and
pected to remove the drug in a process dependent on the
several of the proteins in the four other clusters have been
proton electrochemical gradient. Thus, ofloxacin and chloram-
studied quite intensively (27). However, we are not aware of
phenicol transport into whole cells was examined in the pres-
any study of proteins in the cluster defined by VMAT. In this
ence or absence of the proton uncoupler CCCP. In the pres-
cluster three differentiated branches with sequence similarities
ence of CCCP accumulation levels were very similar in all
and differences can be distinguished: one of the mammalian
strains, representing the equilibration value for the two com-
proteins, one from a member of the Corynebacterineae, and the
pounds (data not shown). In the absence of CCCP, accumula-
third from an archaeon. All of the motifs identified by Paulsen
tion levels differed significantly among the different strains.
et al. (27) are conserved in cluster C and some specific features
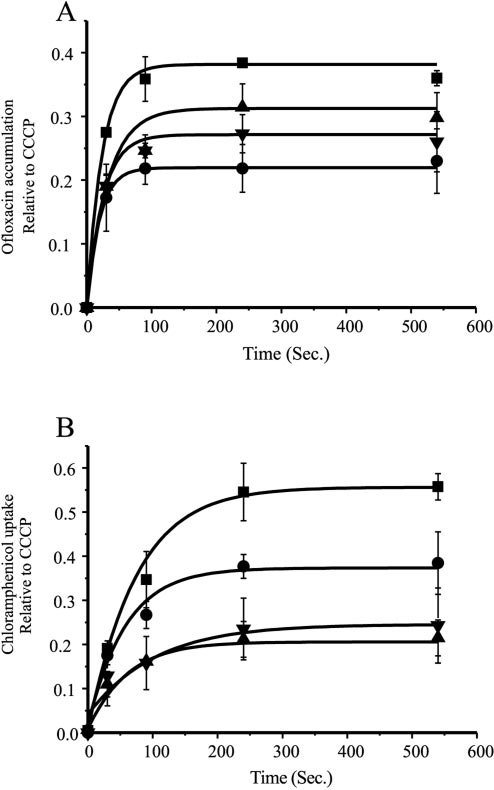
The data in Fig. 4 are presented relative to uptake in the
have been identified here, notably, a putative ion pair in the
presence of CCCP.
membrane domain is hinted by comparison with other mem-
The control strain exhibited the highest uptake for both
bers of the family.
chloramphenicol and ofloxacin. Cells expressing HSmdr accu-
Two putative protein sequences from the Corynebacterineae
mulated ofloxacin twofold less than control. In CGmdr- and
branch, MSmdr from Mycobacterium smegmatis and CGmdr
MSmdr-expressing cells ofloxacin was accumulated to a level of
from Corynebacterium glutamicum, and one from the archaeal
two-thirds of the control. Chloramphenicol accumulation in
branch, HSmdr from Halobacterium salinarum, were cloned
HSmdr-expressing cells was two-thirds of that observed in the
and expressed in E. coli. The three proteins were expressed
control cells while cells expressing MSmdr and CGmdr accu-
and targeted to the membrane and conferred resistance to two
mulated to a level of about one third of the control (Fig. 4B).
quinolones and to chloramphenicol. Although the chloram-
The uptake profile of the novel multidrug transporters is in
phenicol resistance conferred by CGmdr and MSmdr is lower
good accordance with their resistance profile. The result sup-
than that conferred by other multidrug transporters, the quin-
ports the contention that transport catalyzed by the three pro-
olone resistance conferred by the three homologues is in the
teins is driven by proton electrochemical gradient.
same range as that conferred by other DHA12 drug transport-
Protein expression. To characterize the system at the pro-
ers. For example, the ciprofloxacin level that may be endured
tein level the three putative transporters (tagged with Myc-
by MdfA-expressing cells is four times higher than that of the
His) (23) were expressed in E. coli HMS174. To test expression
control (6). The MIC of ofloxacin for E. coli overexpressing
levels, membranes from known volumes of IPTG-induced cul-
NorA is four times higher than for its control (24).
tures were solubilized in denaturing buffer (1% SDS, 6 M urea,
Transport of chloramphenicol and ofloxacin by E. coli
15 mM Tris-Cl, pH 7.2, 150 mM NaCl), purified on Ni⫹2-
JM109 expressing the new transporters was measured in the
nitrilotriacetic acid beads and analyzed by SDS-polyacrylamide
presence and absence of CCCP. The effect of CCCP on trans-
gel electrophoresis (PAGE).
port suggests that, as predicted for an MFS transporter, the
As judged by Coomassie stain of the purified protein, the
process is driven by the proton electrochemical gradient gen-
CLONING OF NOVEL DRUG ANTIPORTERS
FIG. 4. Antibiotic accumulation in E. coli. A—Ofloxacin uptake in
JM109 bearing pT7-7 CGmdr (�), pT7-7 MSmdr (Œ), pT7-7 HSmdr(F), and the control (■). Data represent relative uptake (uptake di-
FIG. 3. Resistance to toxic compounds. The growth of E. coli
vided by the maximum uptake in the presence of CCCP). Ofloxacin
JM109 expressing CGmdr (�), MSmdr (Œ), HSmdr (F), and the con-
was added to 200 l of bacterial culture (OD
⫽ 20) to reach the final
trol (■) in the presence of the indicated concentrations of chloram-
concentration of 30 M. At the indicated times bacteria were sepa-
phenicol (A) or ofloxacin (B). Bacterial growth after 8 h is shown
rated from the medium by rapid filtration and ofloxacin was measured
relative to growth without antibiotics. The experiment was repeated
by fluorescence. CCCP was added to parallel tubes to a final concen-
three times. One representative experiment is shown.
tration of 0.5 mM. B— Chloramphenicol uptake in the same strains ofJM109 relative to uptake after addition of CCCP. Chloramphenicoluptake was measured as described above for ofloxacin except that thelatter was replaced with [3H]chloramphenicol at a final concentration
erated across the cytoplasmic membrane by primary pumps.
of 0.5 M. The experiment was repeated three times. One represen-
Since resistance is also observed at alkaline pH values (data
tative experiment is shown.
not shown) where the only driving force is the membranepotential, the results might suggest that the efflux is an elec-trogenic process involving the exchange of more than one
that conferred by the other two homologues, was accompanied
proton with a substrate molecule (for detailed discussion see
by efficient ofloxacin removal.
references 16 and 31).). There is a good agreement between
Our knowledge of the function of drug transporters in ar-
drug resistance and the efflux activity measured here. CGmdr
chaea and in C. glutamicum and related organisms is very
and MSmdr conferred resistance to both chloramphenicol and
limited. The studies presented here provide the first report of
ofloxacin, and both transporters removed these compounds to
an MFS drug transporter for Halobacterium sp. strain NRC1.
similar levels. A relatively weak resistance to chloramphenicol
In the case of C. glutamicum there is, as far as we know, only
was detected in cells expressing HSmdr and the ability of these
one report of an MFS drug transporter (12). CGmdr is highly
cells to remove chloramphenicol was significantly lower than
similar to two uncharacterized open reading frames in two
that of cells expressing the other transporters. On the other
other actinobacteria, Corynebacterium efficiens (70% identity,
hand the quinolone resistance conferred by HSmdr, similar to
starting at base 1262385) and Nocardia farcinica (40% identity,
VARDY ET AL.
J. BACTERIOL.
Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium
channel: molecular basis of K⫹ conduction and selectivity. Science 280:
69–77.
5. Dutzler, R., E. B. Campbell, M. Cadene, B. T. Chait, and R. MacKinnon.
2002. X-ray structure of a ClC chloride channel at 3.0 A reveals the molec-
ular basis of anion selectivity. Nature 415:287–294.
6. Edgar, R., and E. Bibi. 1997. MdfA, an Escherichia coli multidrug resistance
protein with an extraordinarily broad spectrum of drug recognition. J.
Bacteriol. 179:2274–2280.
7. Goldberg, E. B., T. Arbel, J. Chen, R. Karpel, G. A. Mackie, S. Schuldiner,
and E. Padan. 1987. Characterization of a Na⫹/H⫹ antiporter gene of
Escherichia coli. Proc. Natl. Acad. Sci. USA 84:2615–2619.
8. Gottesman, M. M., I. Pastan, and S. V. Ambudkar. 1996. P-glycoprotein and
multidrug resistance. Curr. Opin. Genet. Dev. 6:610–617.
9. Gutman, N., S. Steiner-Mordoch, and S. Schuldiner. 2003. An amino acid
cluster around the essential Glu-14 is part of the substrate-and proton-
binding domain of EmrE, a multidrug transporter from Escherichia coli.
J. Biol. Chem. 278:16082–16087.
10. Huang, Y., M. J. Lemieux, J. Song, M. Auer, and D. N. Wang. 2003. Structure
and mechanism of the glycerol-3-phosphate transporter from Escherichia
FIG. 5. Expression of the transporters in HMS174 strain. E. coli
coli. Science 301:616–620.
HMS174 cells carrying a pT7-7 plasmid with the gene coding for each
11. Iwaki, S., N. Tamura, T. Kimura-Someya, S. Nada, and A. Yamaguchi. 2000.
of the homologues were used for expression. After harvesting, mem-
Cysteine-scanning mutagenesis of transmembrane segments 4 and 5 of the
branes were prepared as described in Materials and Methods and
Tn10-encoded metal-tetracycline/H⫹ antiporter reveals a permeability bar-
solubilized in 2% SDS–6 M urea. A. Different culture volumes were
rier in the middle of a transmembrane water-filled channel. J. Biol. Chem.
used for protein purification of SDS-Urea solubilized membranes with
Ni-nitrilotriacetic acid beads as described in Materials and Methods.
12. Jager, W., J. Kalinowski, and A. Puhler. 1997. A Corynebacterium glutami-
cum gene conferring multidrug resistance in the heterologous host Esche-
CGmdr was purified from 250 l culture, MSmdr, HSmdr, and the
richia coli. J. Bacteriol. 179:2449–2451.
control from 2.5 ml, and an additional HSmdr preparation from 25 ml
13. Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002.
culture. B. Total membranes were solubilized in denaturing buffer,
Crystal structure and mechanism of a calcium-gated potassium channel.
separated by SDS-PAGE, and underwent Western blot analysis with
rabbit anti-Myc as the primary antibody. The samples shown are from
14. Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B. T. Chait, and R. Mac-
1 l CGmdr, 2 l MSmdr, and 10 l HSmdr membranes (25 mg
Kinnon. 2003. X-ray structure of a voltage-dependent K⫹ channel. Nature
15. Kaback, H. R. 1971. Bacterial membranes. Methods Enzymol. 22:99–120.
16. Lewinson, O., J. Adler, G. J. Poelarends, P. Mazurkiewicz, A. J. Driessen,
and E. Bibi. 2003. The Escherichia coli multidrug transporter MdfA catalyzes
starting at base 4621748). Expression in the natural hosts was
both electrogenic and electroneutral transport reactions. Proc. Natl. Acad.
not studied here, but the existence of close homologues in
Sci. USA 100:1667–1672.
17. Li, X. Z., L. Zhang, and H. Nikaido. 2004. Efflux pump-mediated intrinsic
other bacteria hints of a natural role which may involve mul-
drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Che-
tidrug transporter activity.
Drug resistance in mycobacteria has been studied quite ex-
18. Liu, Y., D. Peter, A. Roghani, S. Schuldiner, G. G. Prive, D. Eisenberg, N.
Brecha, and R. H. Edwards. 1992. A cDNA that suppresses MPP⫹ toxicity
tensively because of the fact that even now in the 21st century,
encodes a vesicular amine transporter. Cell 70:539–551.
Mycobacterium tuberculosis is still the deadliest single killer
19. Marger, M. D., and M. H. Saier, Jr. 1993. A major superfamily of trans-
according to a World Health Organization report (39). Myco-
membrane facilitators that catalyse uniport, symport and antiport. Trends
Biochem. Sci. 18:13–20.
bacteria are gram-positive bacteria intrinsically resistant to a
20. Merickel, A., H. R. Kaback, and R. H. Edwards. 1997. Charged residues in
variety of antibacterials. Several mycobacterial MFS multi-
transmembrane domains II and XI of a vesicular monoamine transporter
drug transporters have been reported in M. tuberculosis and
form a charge pair that promotes high affinity substrate recognition. J. Biol.
Chem. 272:5403–5408.
M. smegmatis (17). Inactivation of chromosomal genes in
21. Miroux, B., and J. E. Walker. 1996. Overproduction of proteins in Esche-
M. smegmatis points to several transporters that may mediate
richia coli: mutant hosts that allow synthesis of some membrane proteins and
globular proteins at high levels. J. Mol. Biol. 260:289–298.
the intrinsic drug resistance (17). The M. smegmatis trans-
22. Mortimer, P. G., and L. J. Piddock. 1991. A comparison of methods used for
porter characterized here is a homologue (50% identity) of an
measuring the accumulation of quinolones by Enterobacteriaceae, Pseudo-
open reading frame in M. avium (unfinished genome, starting
monas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother.
28:639–653.
at base 5080617).
23. Muth, T. R., and S. Schuldiner. 2000. A membrane-embedded glutamate is
required for ligand binding to the multidrug transporter EmrE. EMBO J.
24. Ng, E. Y., M. Trucksis, and D. C. Hooper. 1994. Quinolone resistance
This work was supported by grant 2003-309 from the United States-
mediated by norA: physiologic characterization and relationship to flqB, a
Israel Binational Science Foundation, Jerusalem, Israel, and from
quinolone resistance locus on the Staphylococcus aureus chromosome. An-
the Center for Innovation in Membrane Protein Production (P50
timicrob. Agents Chemother. 38:1345–1355.
25. Nicholas, K. B., H. B. Nicholas, Jr, and D. W. Deerfield. 1997. GeneDoc:
GM73210). Shimon Schuldiner is Mathilda Marks-Kennedy Professor
analysis and visualization of genetic variation. EMBnet News Online 4.
of Biochemistry at the Hebrew University of Jerusalem.
26. Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability
barriers and active efflux. Science 264:382–388.
1. Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback, and S. Iwata.
27. Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent
2003. Structure and mechanism of the lactose permease of Escherichia coli.
multidrug efflux systems. Microbiol. Rev. 60:575–608.
28. Paulsen, I. T., and R. A. Skurray. 1993. Topology, structure and evolution of
2. Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller,
two families of proteins involved in antibiotic and antiseptic resistance in
and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation
eukaryotes and prokaryotes–an analysis. Gene 124:1–11.
of protein database search programs. Nucleic Acids Res. 25:3389–3402.
29. Perriere, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system
3. Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of
for biological sequence banks. Biochimie 78:364–369.
microgram quantities of protein utilizing the principle of protein-dye bind-
30. Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties
ing. Anal. Biochem. 72:248–254.
of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672–693.
4. Doyle, D. A., J. Morais Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L.
31. Rotem, D., and S. Schuldiner. 2004. EmrE, a multidrug transporter from
CLONING OF NOVEL DRUG ANTIPORTERS
Escherichia coli, transports monovalent and divalent substrates with the same
37. Vardy, E., I. T. Arkin, K. E. Gottschalk, H. R. Kaback, and S. Schuldiner.
stoichiometry. J. Biol. Chem. 279:48787–48793.
2004. Structural conservation in the major facilitator superfamily as revealed
32. Saier, M. H., Jr., and I. T. Paulsen. 2001. Phylogeny of multidrug transport-
by comparative modeling. Protein Sci. 13:1832–1840.
ers. Semin. Cell Dev. Biol. 12:205–213.
38. Viklund, H., and A. Elofsson. 2004. Best alpha-helical transmembrane pro-
33. Sambrook, J., R. D. 2001. Molecular cloning: a laboratory manual, 3rd ed.,
tein topology predictions are achieved using hidden Markov models and
vol. 1–3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor,. NY.
evolutionary information. Protein Sci. 13:1908–1917.
34. Schuldiner, S., A. Shirvan, and M. Linial. 1995. Vesicular neurotransmitter
39. World Health Organization. 2005. Global tuberculosis control—surveil-
transporters: from bacteria to humans. Physiol. Rev. 75:369–392.
lance, planning, financing (W.H.O./HTM/TB/2005.349). World Health Or-
35. Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymer-
ganization, Geneva, Switzerland.
ase/promoter system for controlled exclusive expression of specific genes.
40. Yanisch-Perron, C., J. Viera, and J. Messing. 1985. Improved M13 phage
Proc. Natl. Acad. Sci. USA 82:1074–1078.
cloning vectors and host strains: nucleotide sequences of the M13mp18 and
36. Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W:
pUC19 vectors. Gene 33:103–119.
improving the sensitivity of progressive multiple sequence alignment through
41. Yelin, R., and S. Schuldiner. 1995. The pharmacological profile of the ve-
sequence weighting, position-specific gap penalties and weight matrix choice.
sicular monoamine transporter resembles that of multidrug transporters.
Nucleic Acids Res. 22:4673–4680.
FEBS Lett. 377:201–207.
Source: http://biolchem.huji.ac.il/shimons/152%20VMAT_Homologues.pdf
Blackburn et al. Research Involvement and Engagement (2016) 2:5 DOI 10.1186/s40900-016-0019-x Patient-reported quality indicators forosteoarthritis: a patient and publicgenerated self-report measure for primarycare Steven Blackburn1*, Adele Higginbottom1, Robert Taylor2, Jo Bird2, Nina Østerås3, Kåre Birger Hagen3,John J. Edwards1, Kelvin P. Jordan1, Clare Jinks1 and Krysia Dziedzic1
PEDRO CÉSAR CANTÚ MARTÍNEZ* Medio ambiente y salud: un enfoque ecosistémico El concepto ecología, establecida en una interrelación con el resto de los seres vivos y su 1869, por Ernest Haeckel, un natu- hábitat; asimismo, con la estructura y funcionamiento ralista alemán, reconocido como pa- de los ecosistemas en general que lo acogen. De manera dre de la ecología, determina que es








